In-depth use of ScrubletR
2024-02-02
InDepth.RmdInstalling scrubletR
You will need to have the devtools package installed…
devtools::install_github("furlan-lab/scrubletR")Load data
Let’s read in a 10X dataset where T cells from two donors were combined before capture. We then use souporcell to detect genetic demultiplets. Clustering in higher dimension actually works better for visualizing the SNP clustering… First we read in the pca-transformed probability scores from souporcell algorithm which enables us to visualize the two expected genotypes but having set k=3 enables 3 dimensions of data from the algorithm (the first dimension is ‘cell size’ aka UMI count per cell). Therefore we visualize the embedding using the 2nd and 3rd PCs, but label the cells according to their label setting k=2. The following code loads the 10X data, the souporcell data and makes the plot.
rm(list = ls())
suppressPackageStartupMessages({
library(viewmastR)
library(Seurat)
library(scCustomize)
library(scrubletR)
library(ggplot2)
library(magrittr)
})
if(grepl("^gizmo", Sys.info()["nodename"])){
ROOT_DIR2<-"/fh/fast/furlan_s/grp/data/experiments/MM_CarT_5p_vdjt/data/SNF"
} else {
ROOT_DIR2<-"/Users/sfurlan/Library/CloudStorage/OneDrive-SharedLibraries-FredHutchinsonCancerCenter/Furlan_Lab - General/experiments/MM_CART/SNF"
}
counts<-Read10X_h5(file.path(ROOT_DIR2, "sample_filtered_feature_bc_matrix.h5"))
colnames(counts$`Gene Expression`)<-paste0("JP_709_707_IP_", colnames(counts$`Gene Expression`))
seu<-CreateSeuratObject(counts$`Gene Expression`)
scdata<-data.table::fread(file.path(ROOT_DIR2, "clusters_3.tsv"))
seu<-seu[,Cells(seu) %in% scdata$barcode]
seu<-add_souporcell_seurat(seu, file.path(ROOT_DIR2, "clusters_3.tsv"))
scdata<-data.table::fread(file.path(ROOT_DIR2, "clusters_2.tsv"))
seu$geno_from2<-scdata$assignment[match(Cells(seu), scdata$barcode)]
seu$geno_from2[grepl("\\/", seu$geno_from2)]<-"Multiplet"
DimPlot(seu, group.by = "geno_from2", dims = c(2,3), cols = c("black", "grey", "red"))+ggtitle("Souporcell data (overclustered)")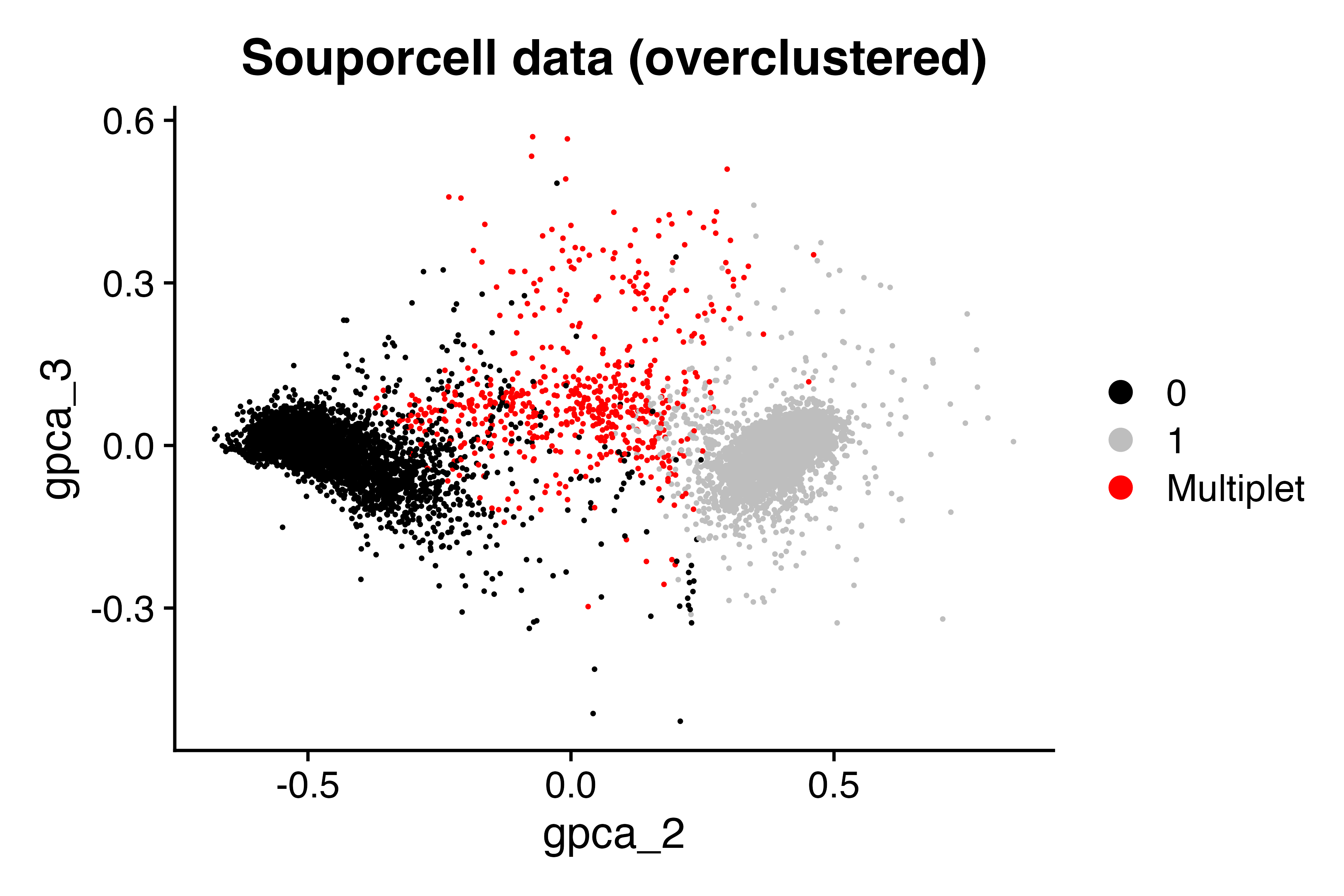
Calculate doublet rate
##
## 0 1 Multiplet
## 0.4387415 0.4950052 0.0662533Looks to be 6.6%. This is what we would expect from a 10X capture of this size.
Run scrublet
seu<-scrublet(seu, min_gene_variability_pctl = 95)
seu$log_ds<-log(seu$doublet_scores)
VlnPlot_scCustom(seu, features = "log_ds", group.by = "geno_from2", pt.size = 0)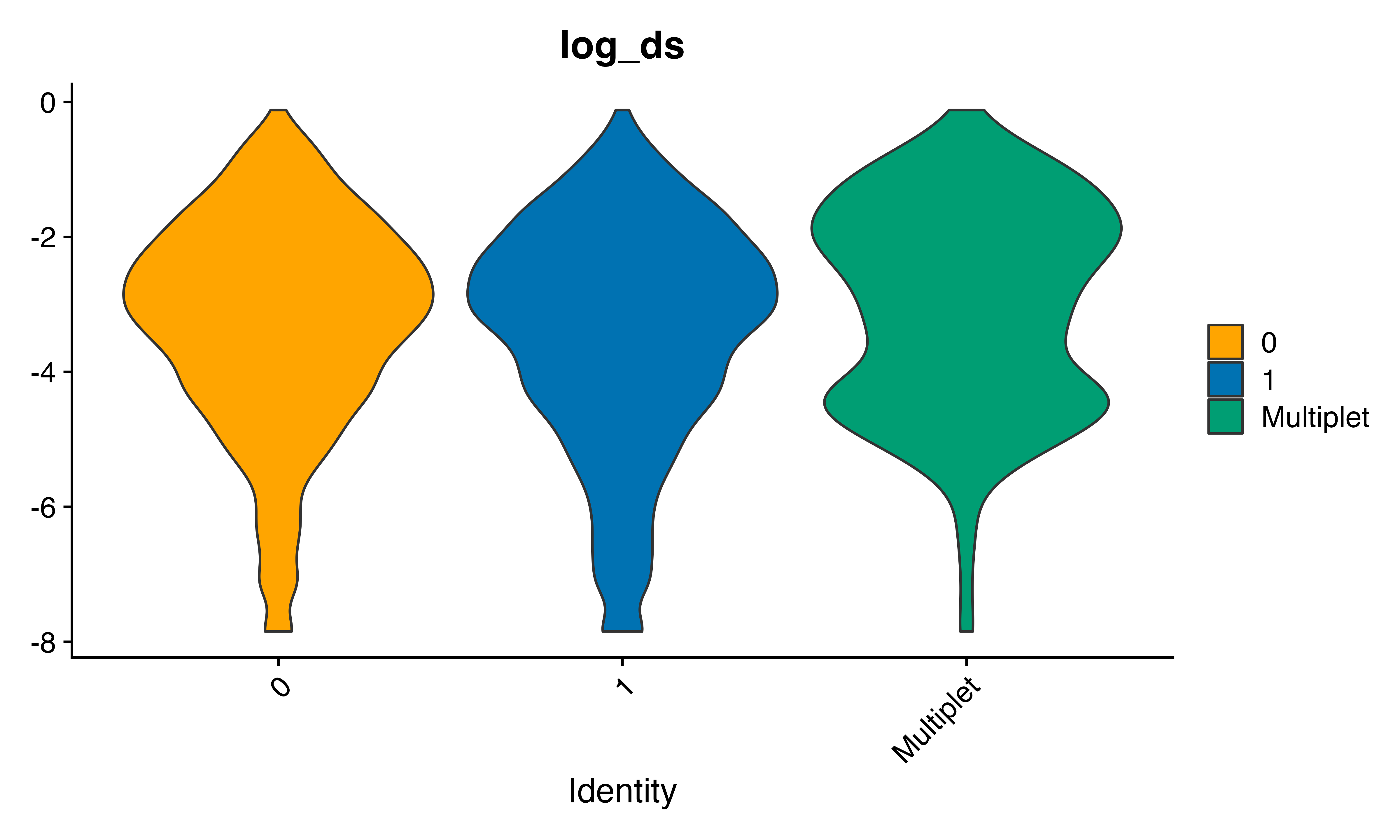
This looks okay. The genetic multiplets have ~ 2 fold increase in scrublet doublet score than cells from each genotype. This is of course not the whole story as we would expect that more than one cell from the same donor could end up in the same droplet. Therefore > 6 percent of the cells in ‘0’ and ‘1’ genotypes above will also be multiplets. Let’s set a threshold and see how the percents fall out.
thresh<-(-2.3) #set by eye
seu$SFdoublet<-seu$log_ds>(thresh)
table(seu$SFdoublet, seu$geno_from2)##
## 0 1 Multiplet
## FALSE 2744 3168 346
## TRUE 1077 1143 231
VlnPlot_scCustom(seu, features = "log_ds", group.by = "geno_from2", pt.size = 0)+geom_hline(yintercept = thresh)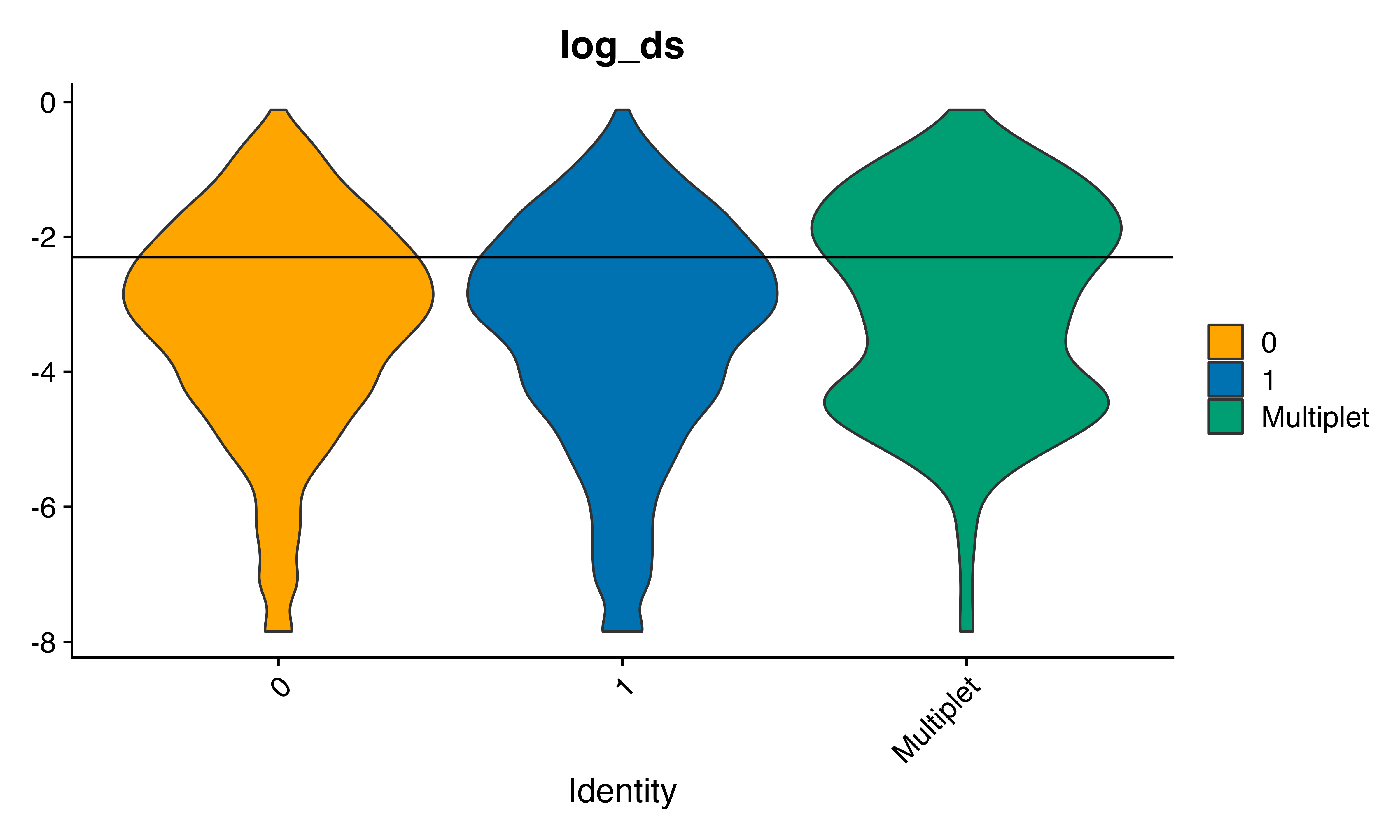
Visualize
Let’s visualize, subset, and re-embed. But we need to fix subsetting in Seurat (seurat 5 is broken)
DefaultAssay(seu)<-"RNA"
seu <-NormalizeData(seu) %>% FindVariableFeatures(nfeatures = 3000) %>% ScaleData() %>% RunPCA(npcs = 50)
#ElbowPlot(seu, 50)
seu<- FindNeighbors(seu, dims = 1:30) %>% FindClusters() %>% RunUMAP(dims = 1:30)## Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
##
## Number of nodes: 8709
## Number of edges: 354588
##
## Running Louvain algorithm...
## Maximum modularity in 10 random starts: 0.8566
## Number of communities: 13
## Elapsed time: 0 seconds
DimPlot(seu)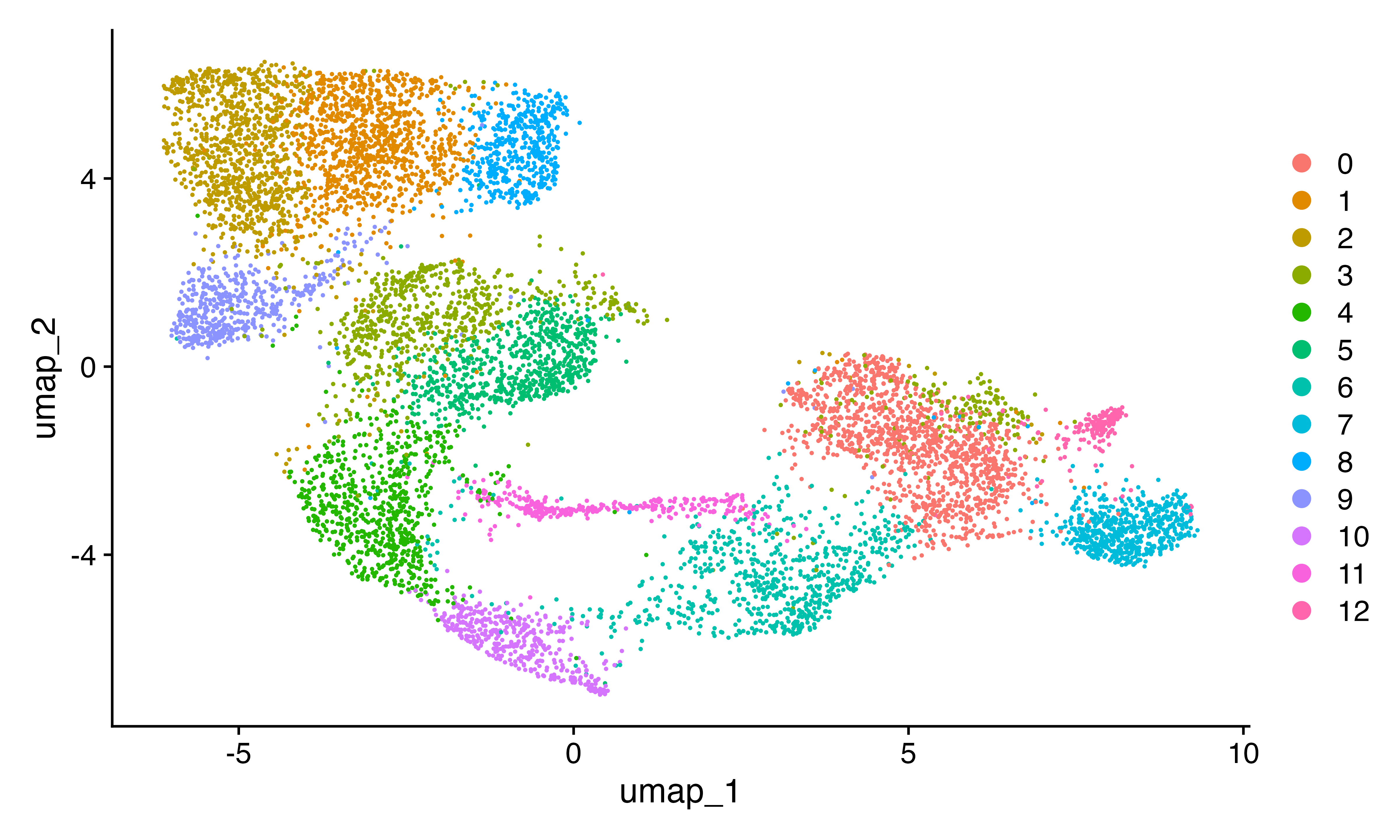
FeaturePlot_scCustom(seu, features = "CD4")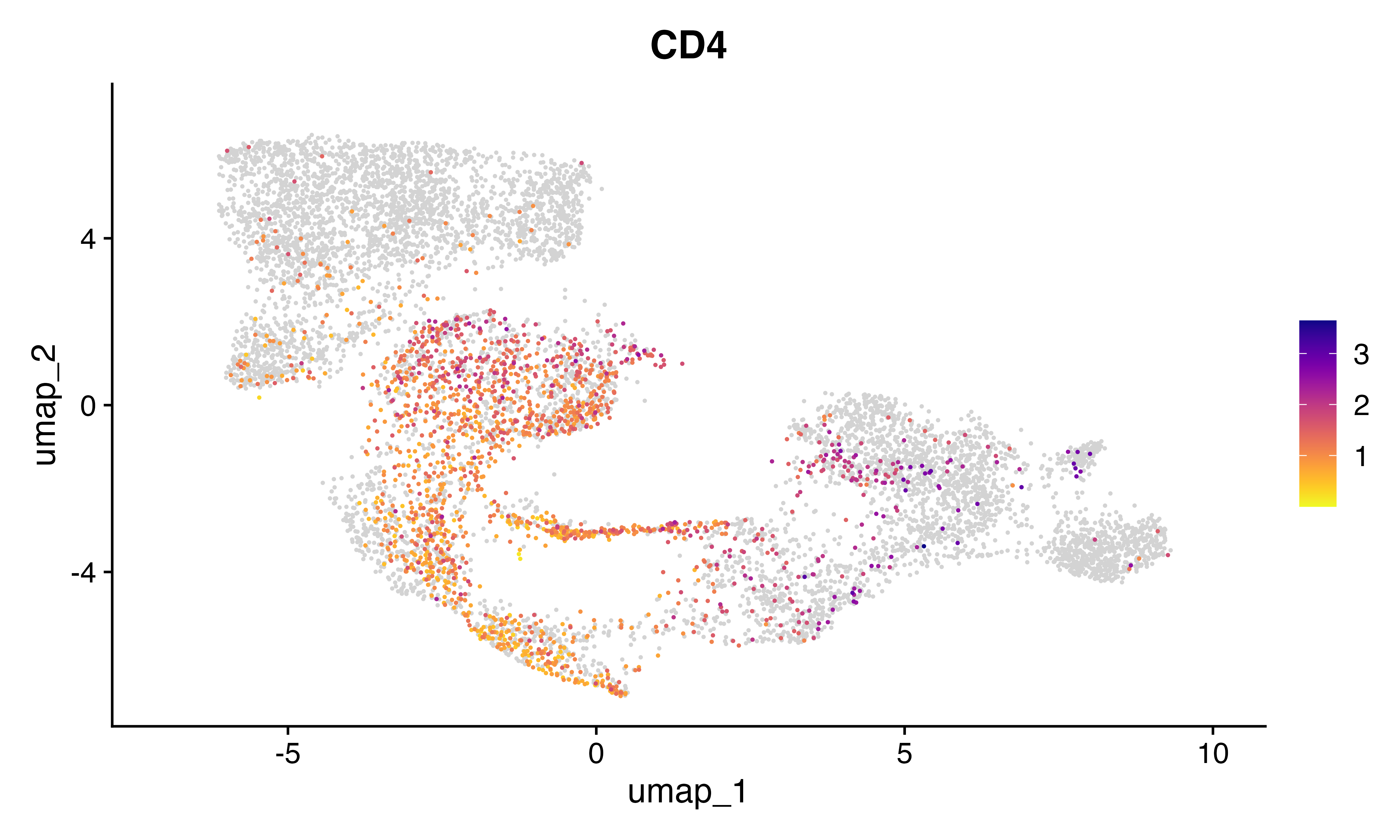
FeaturePlot_scCustom(seu, features = "CD8A")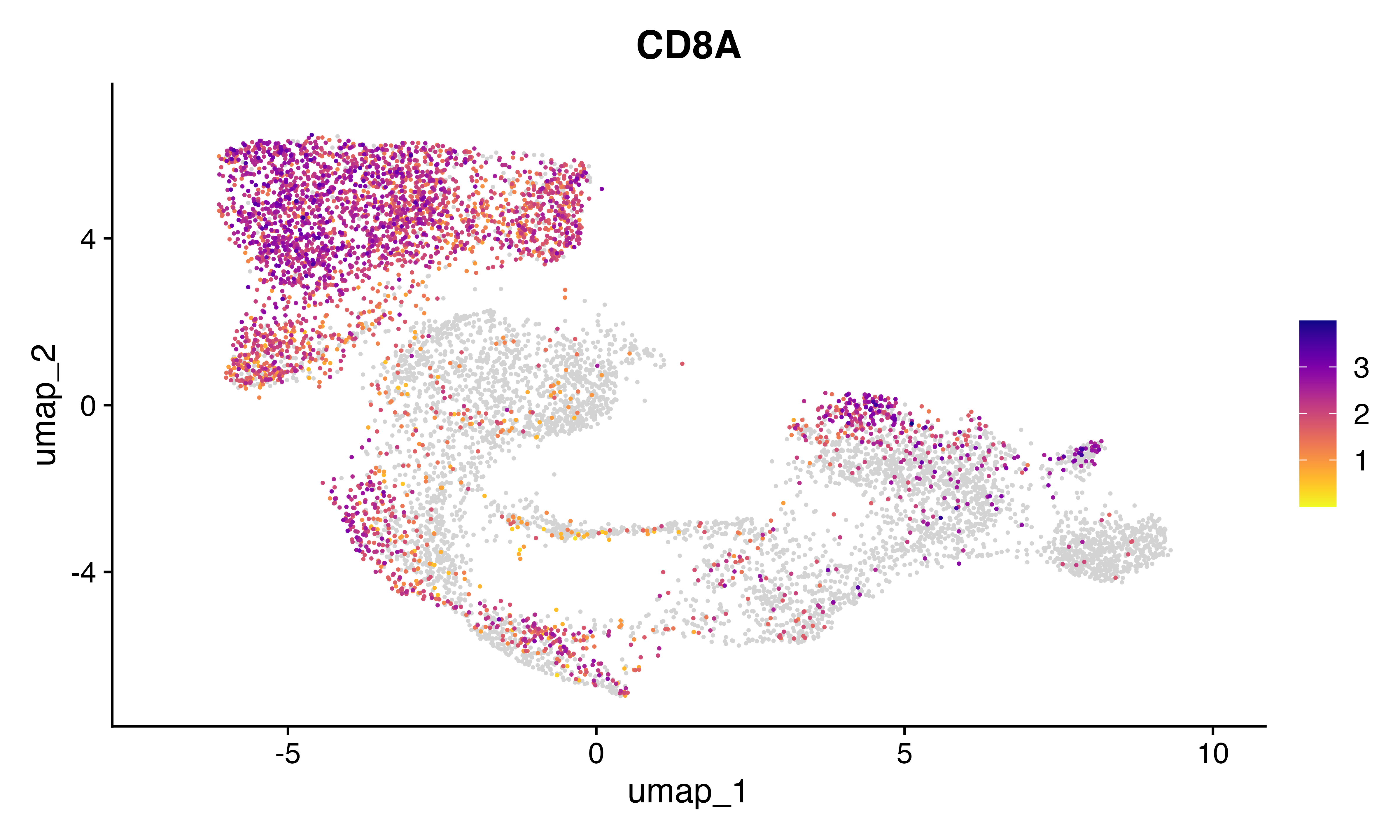
FeaturePlot_scCustom(seu, features = "doublet_scores")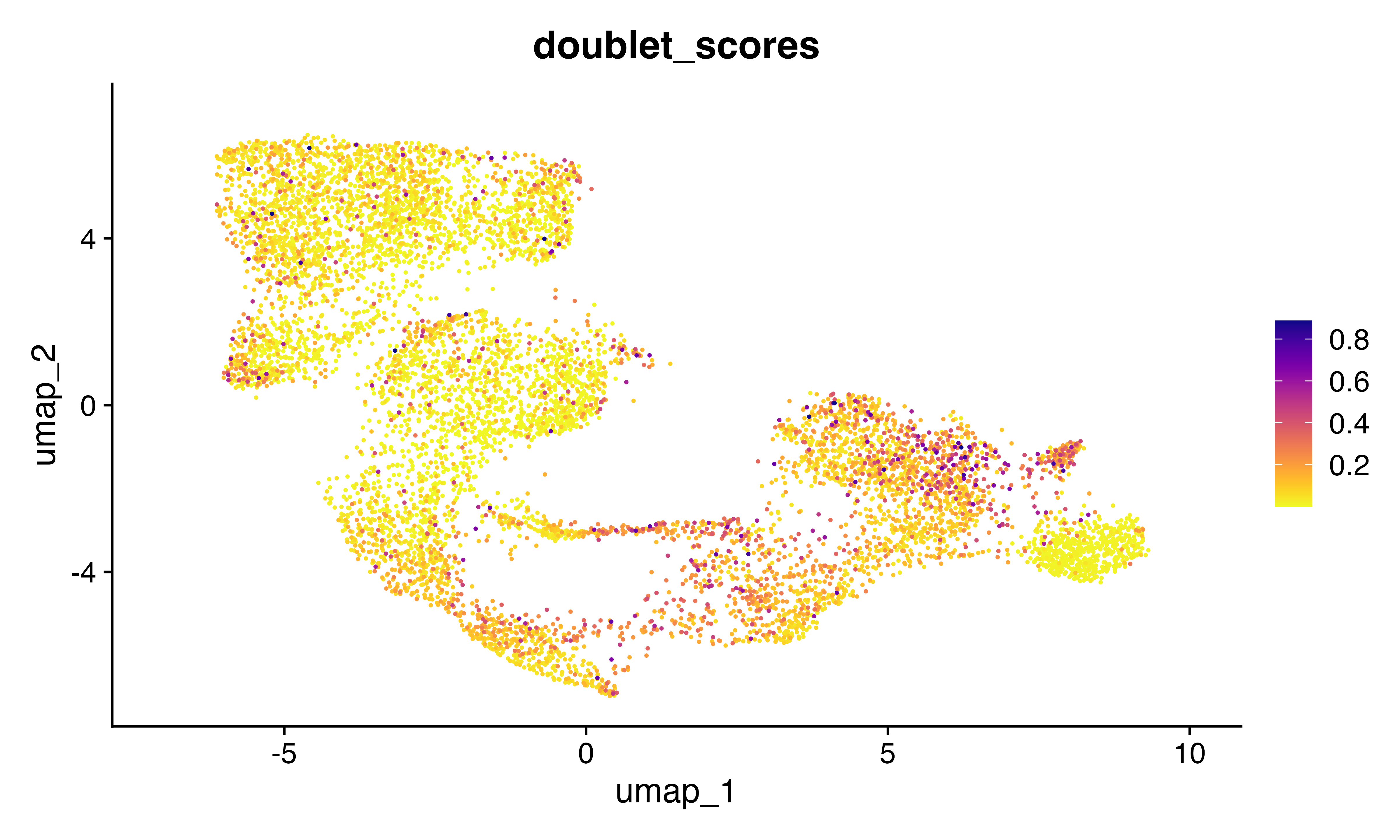
FeaturePlot_scCustom(seu, features = "log_ds")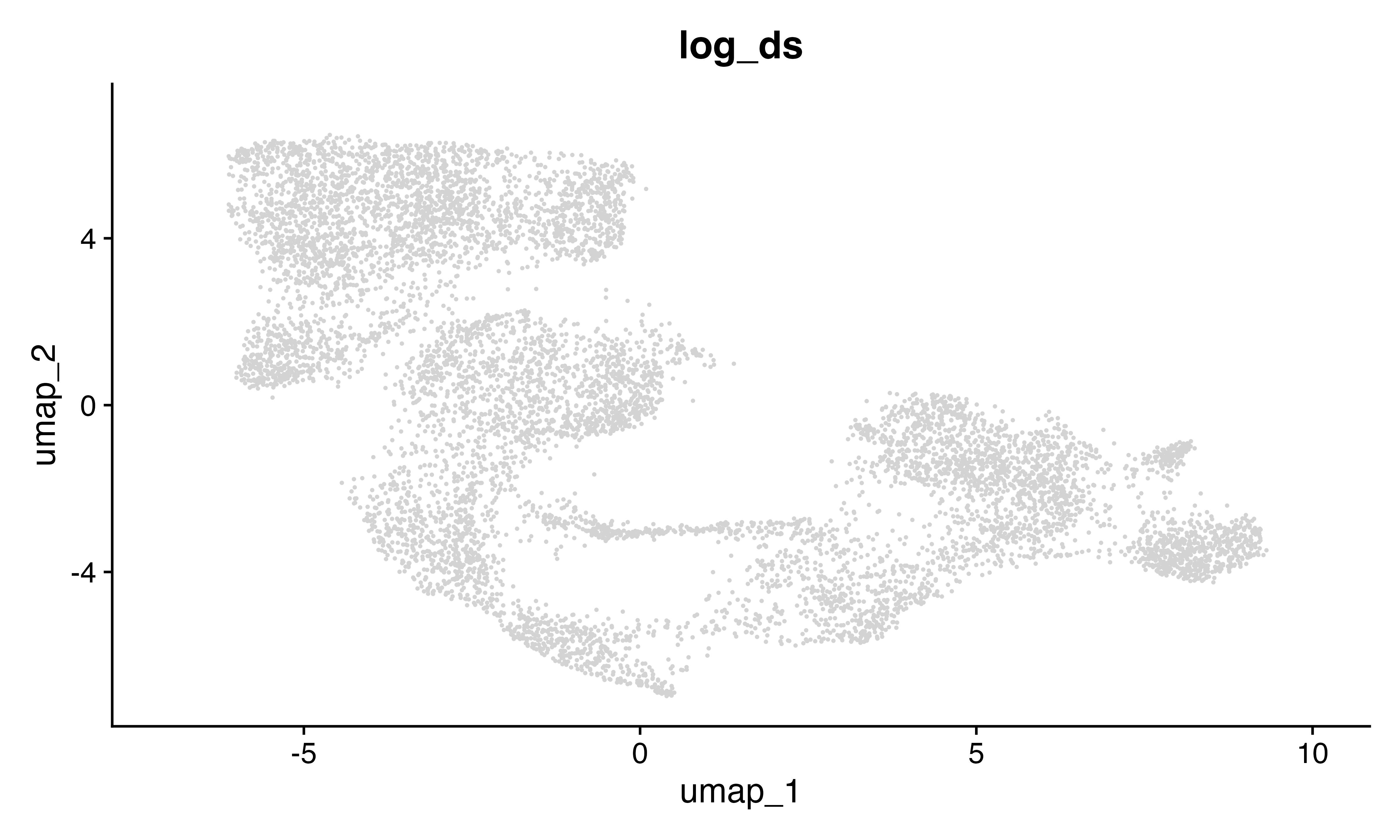
DimPlot(seu, group.by = "predicted_doublets")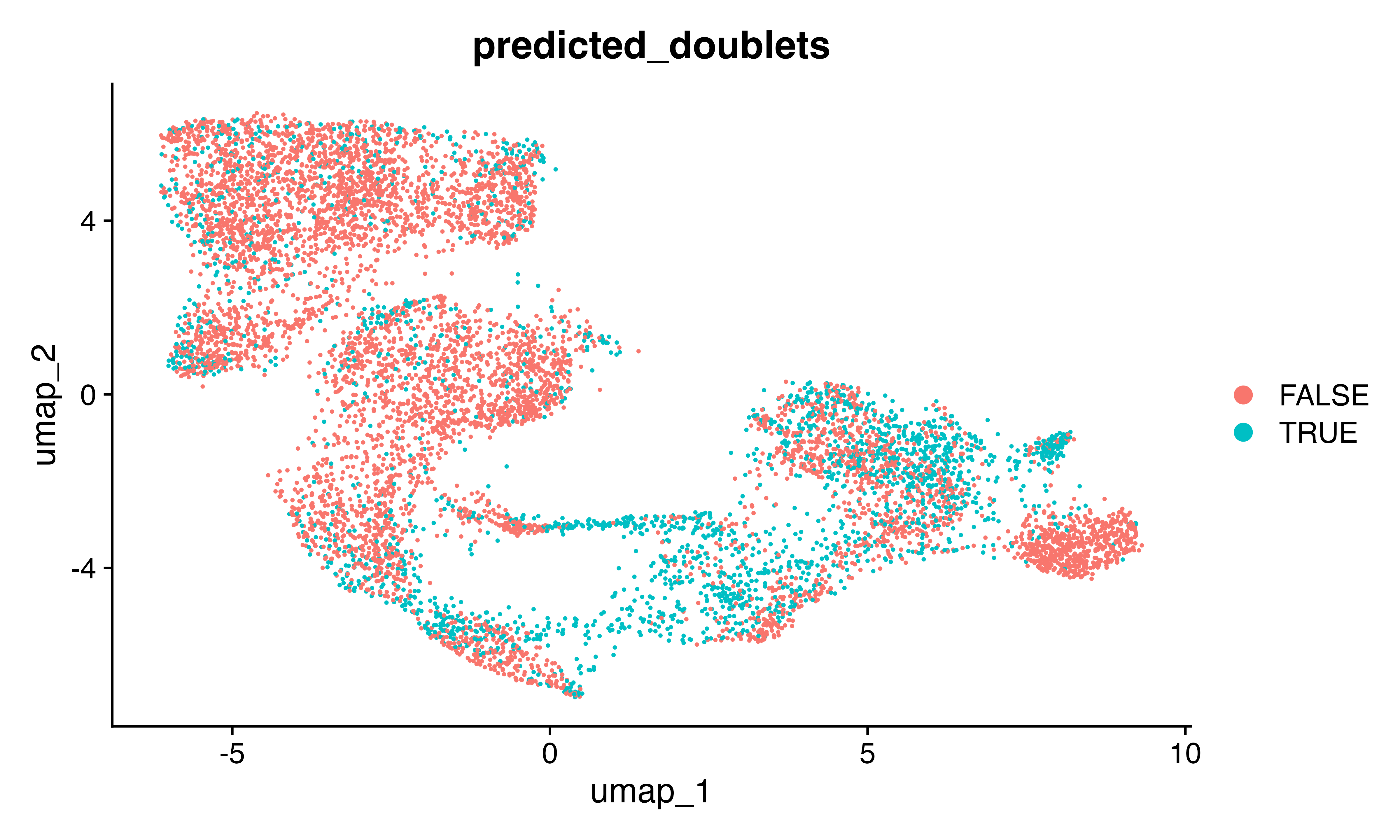
DimPlot(seu, group.by = "SFdoublet")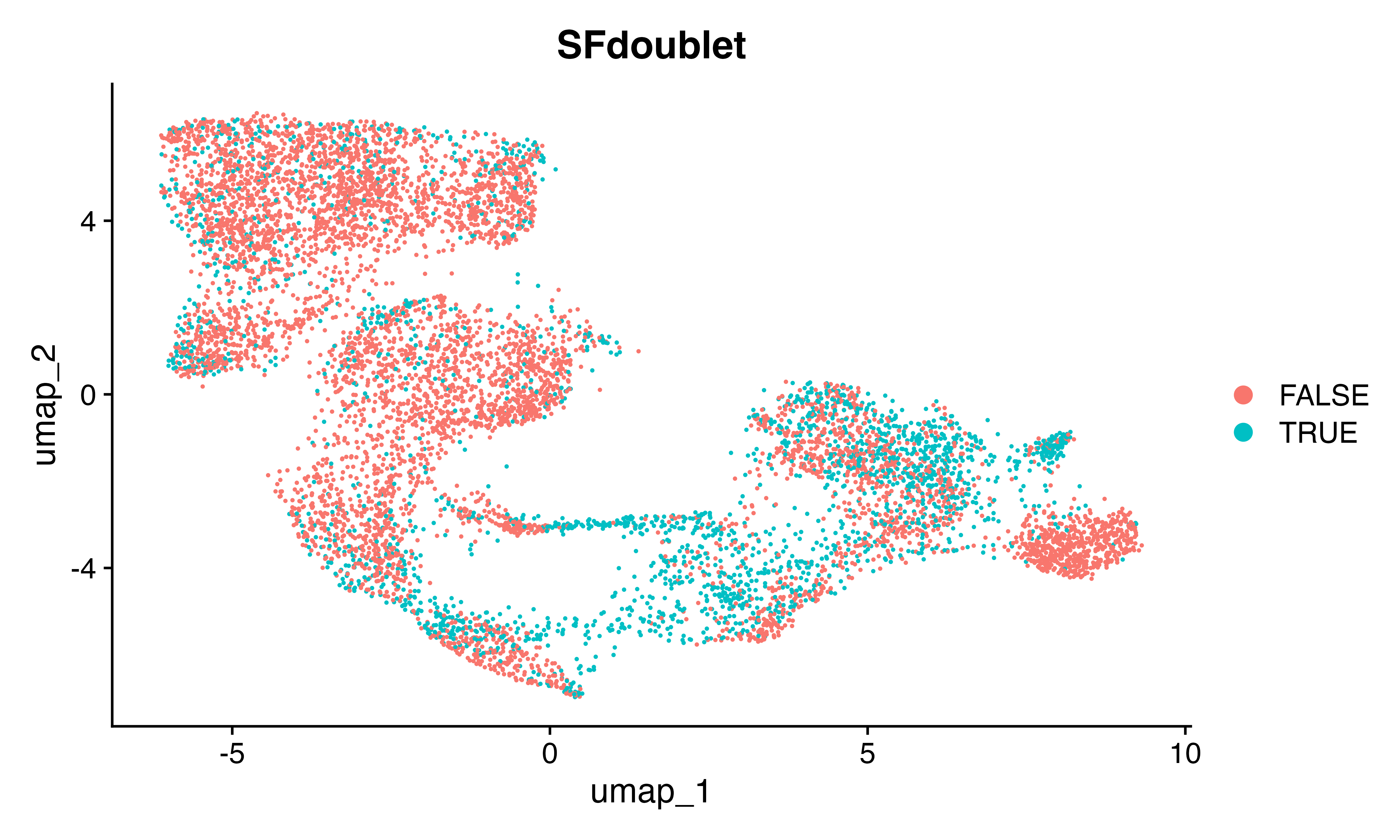
DimPlot(seu, group.by = "geno_from2")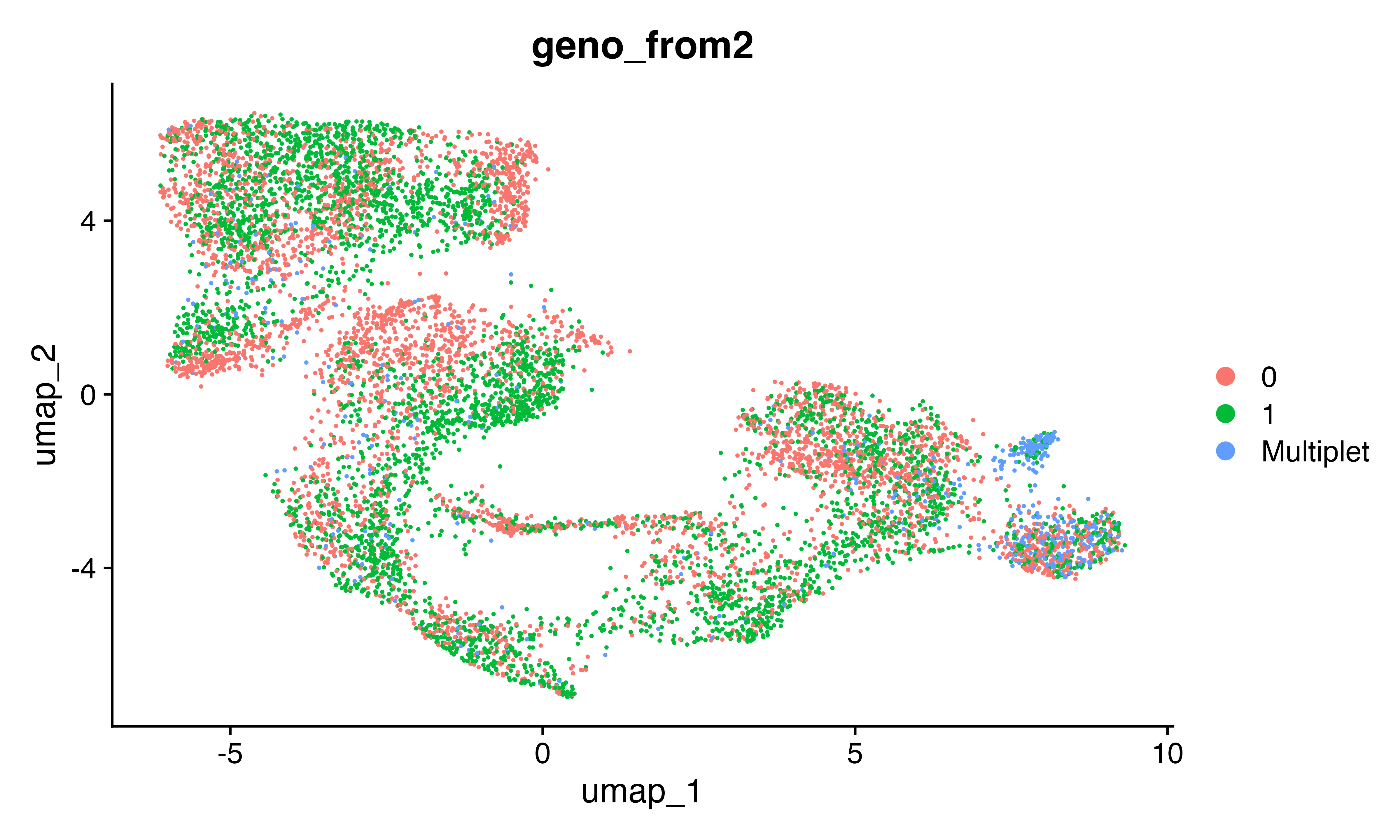
confusion_matrix(factor(seu$SFdoublet), factor(seu$predicted_doublets)) #pretty good!!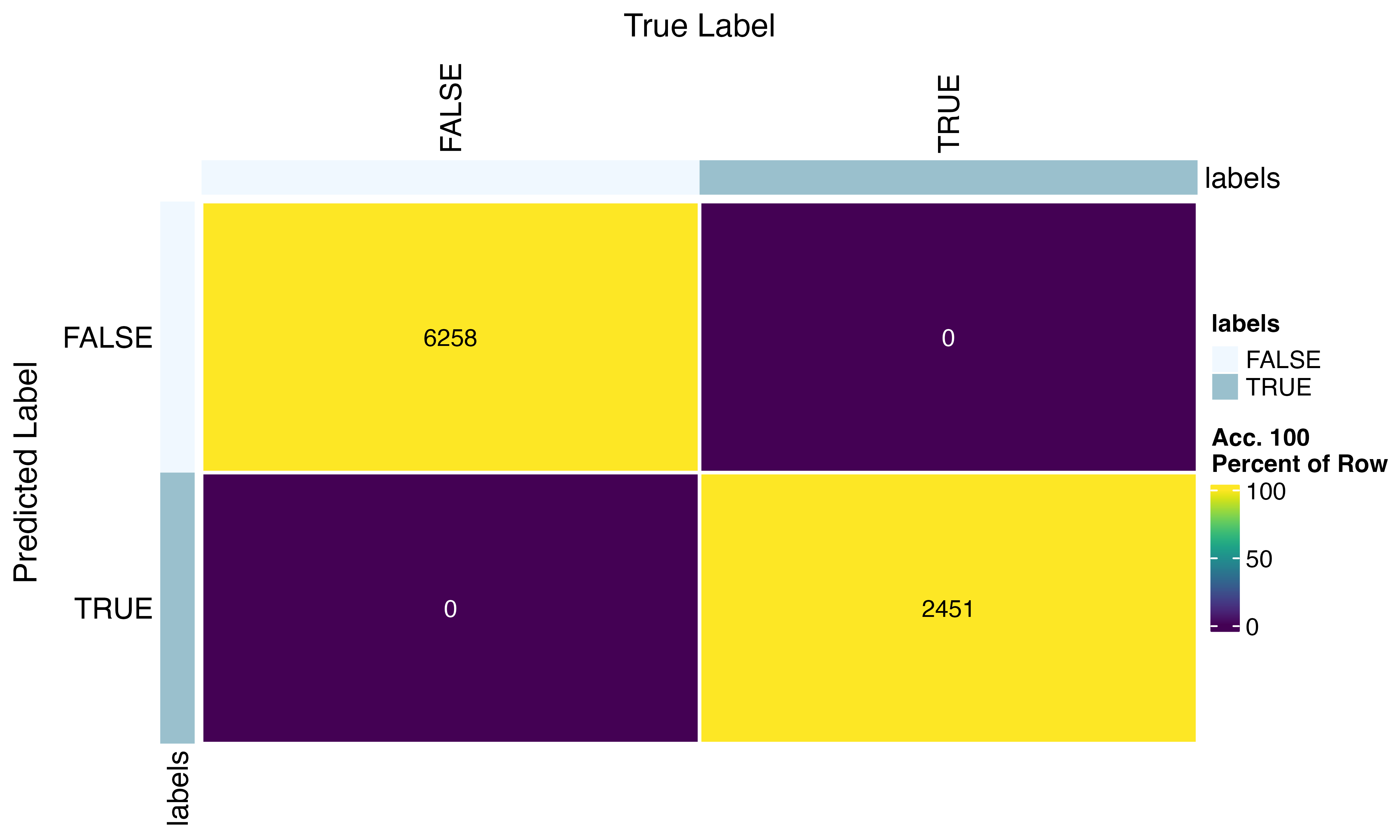
seu$cell_bool<-!seu$SFdoublet & seu$geno_from2 != "Multiplet"
table(seu$cell_bool)##
## FALSE TRUE
## 2797 5912
DimPlot(seu, group.by = "cell_bool")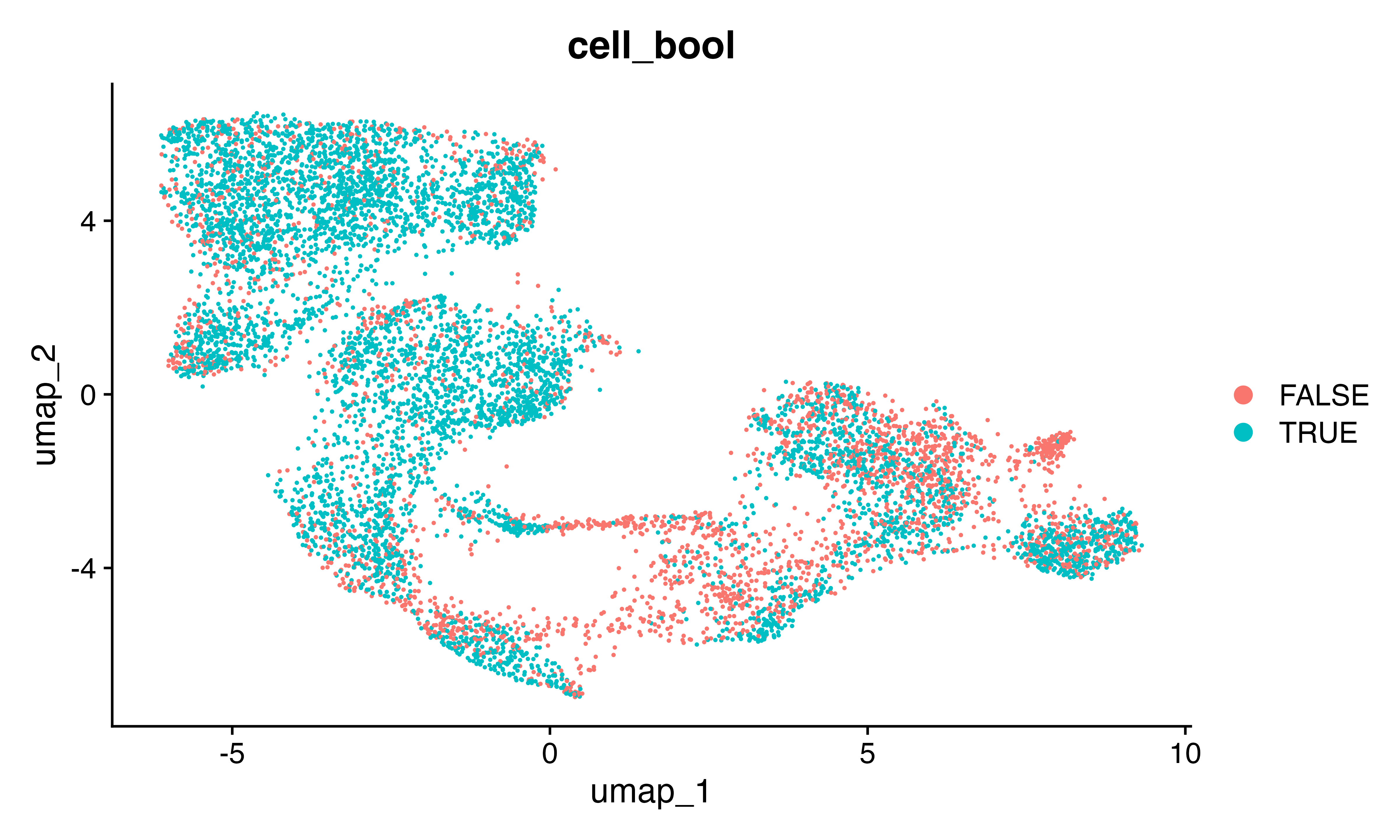
subset_cells_seurat<-function(seu, cell_bool){
if(!length(cell_bool)==length(Cells(seu))){stop("cell_bool argument length does not match the number of cells in Seurat object")}
assays<-Assays(seu)
suppressWarnings(layers<-lapply(assays, function(assay) {
subset(GetAssay(seu, assay = assay), Cells(seu)[cell_bool])
}))
newseu<-CreateSeuratObject(layers[[1]], meta.data = seu@meta.data[cell_bool,])
for(i in 2:length(assays)){
newseu[[assays[i]]]<-layers[[i]]
}
newseu
}
seu<-subset_cells_seurat(seu, seu$cell_bool)
seu <-NormalizeData(seu) %>% FindVariableFeatures(nfeatures = 3000) %>% ScaleData() %>% RunPCA(npcs = 50)
#ElbowPlot(seu, 50)
seu<- invisible(FindNeighbors(seu, dims = 1:30) %>% FindClusters() %>% RunUMAP(dims = 1:30))## Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
##
## Number of nodes: 5912
## Number of edges: 253283
##
## Running Louvain algorithm...
## Maximum modularity in 10 random starts: 0.8540
## Number of communities: 12
## Elapsed time: 0 seconds
DimPlot(seu)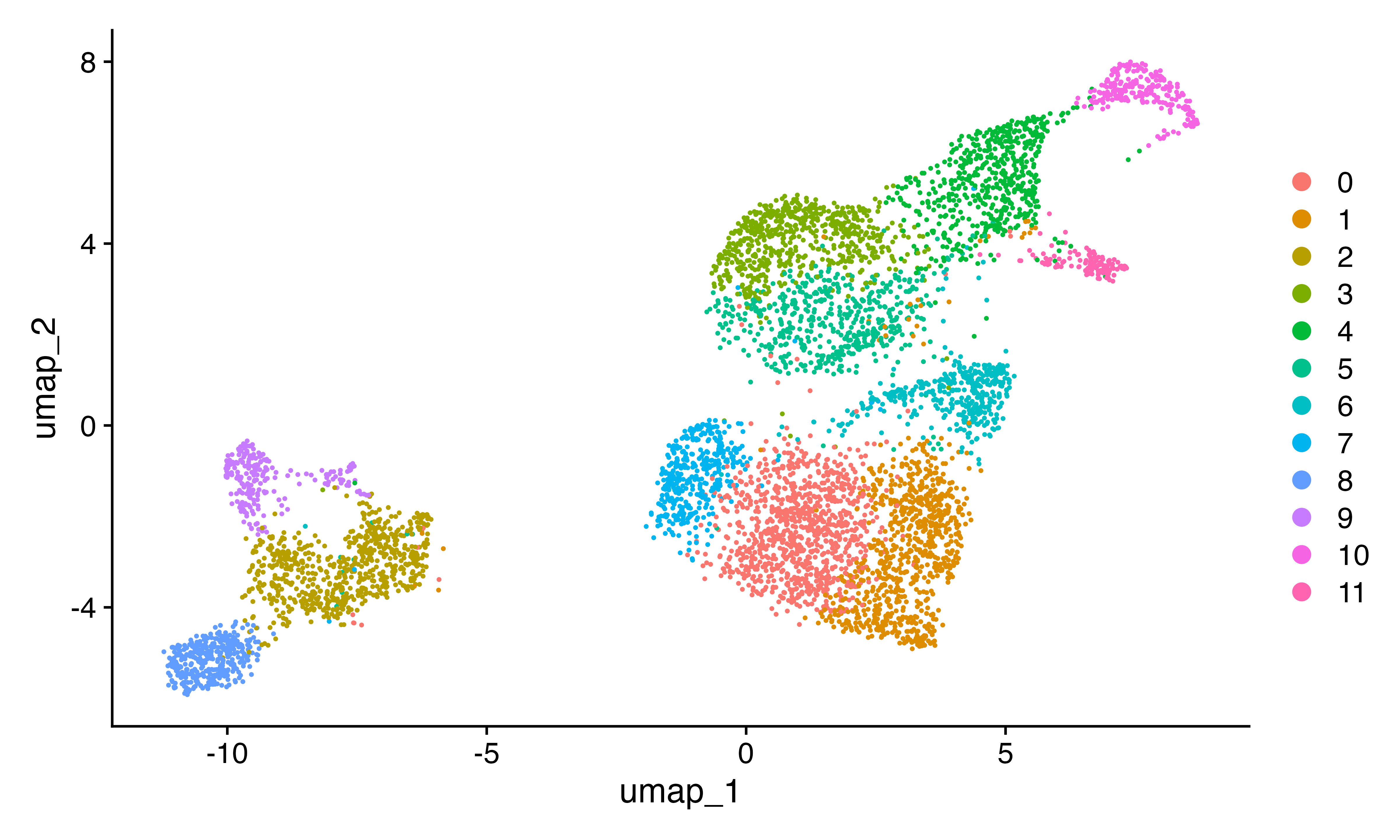
FeaturePlot_scCustom(seu, features = "doublet_scores")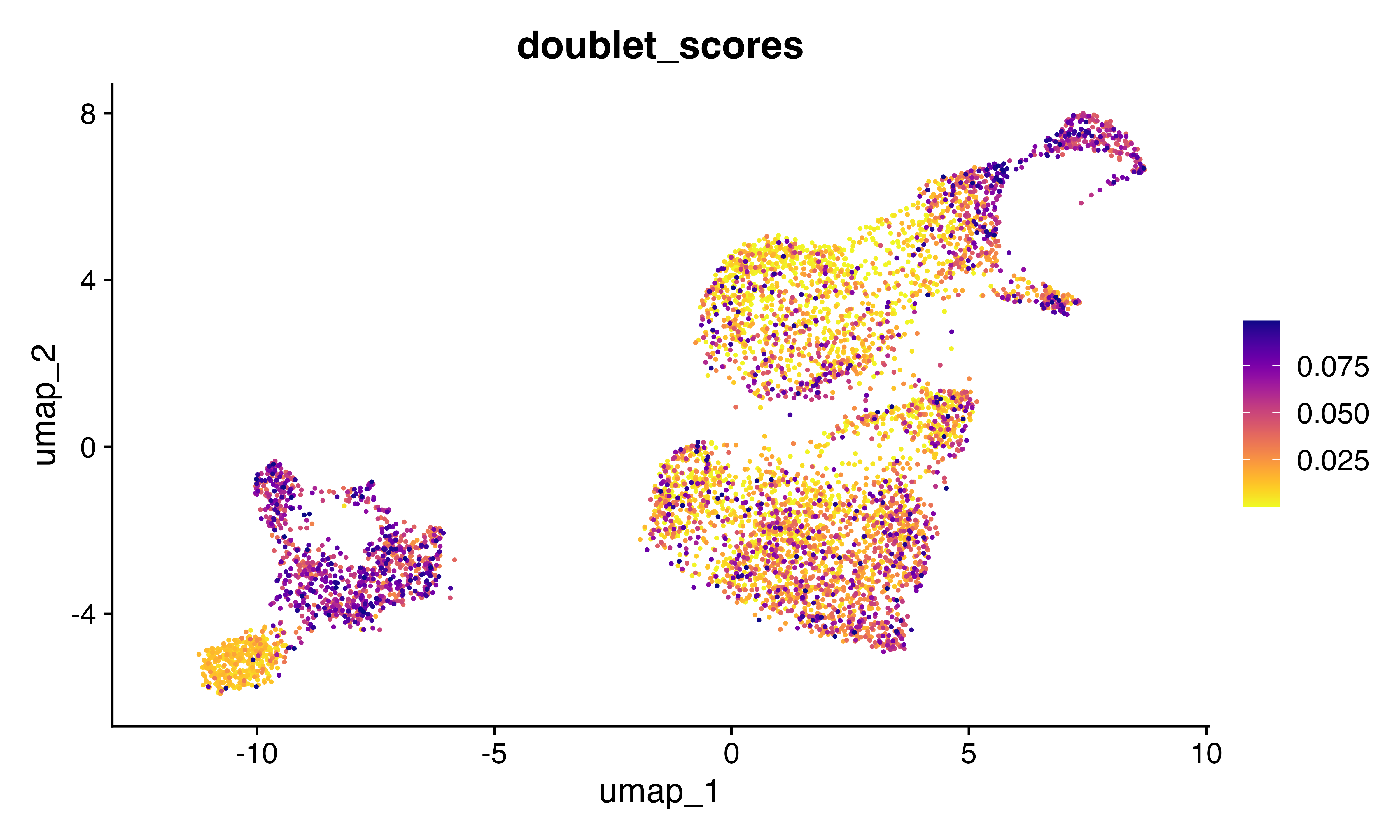
Rerun scrublet
seu<-scrublet(seu)
FeaturePlot_scCustom(seu, features = "doublet_scores")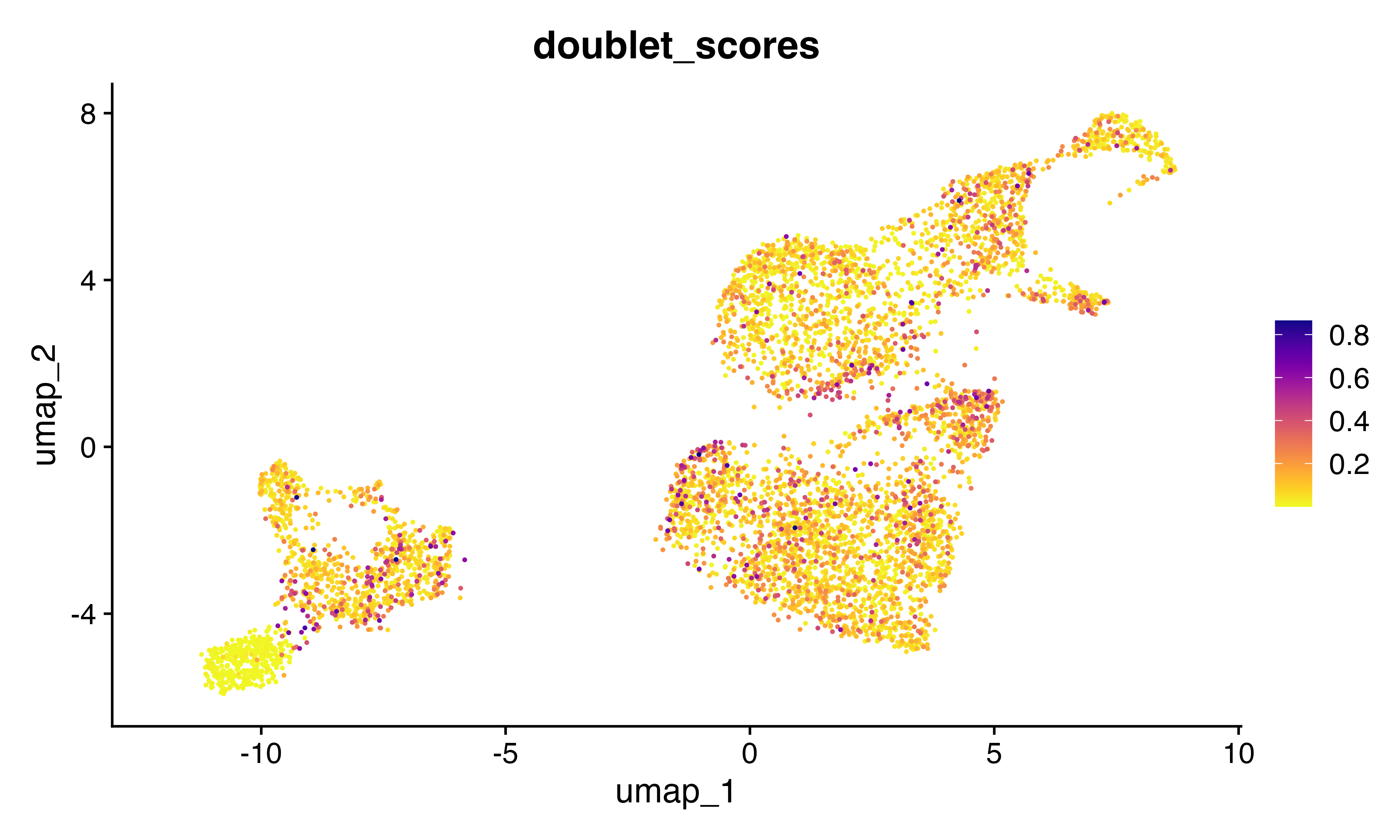
DimPlot(seu)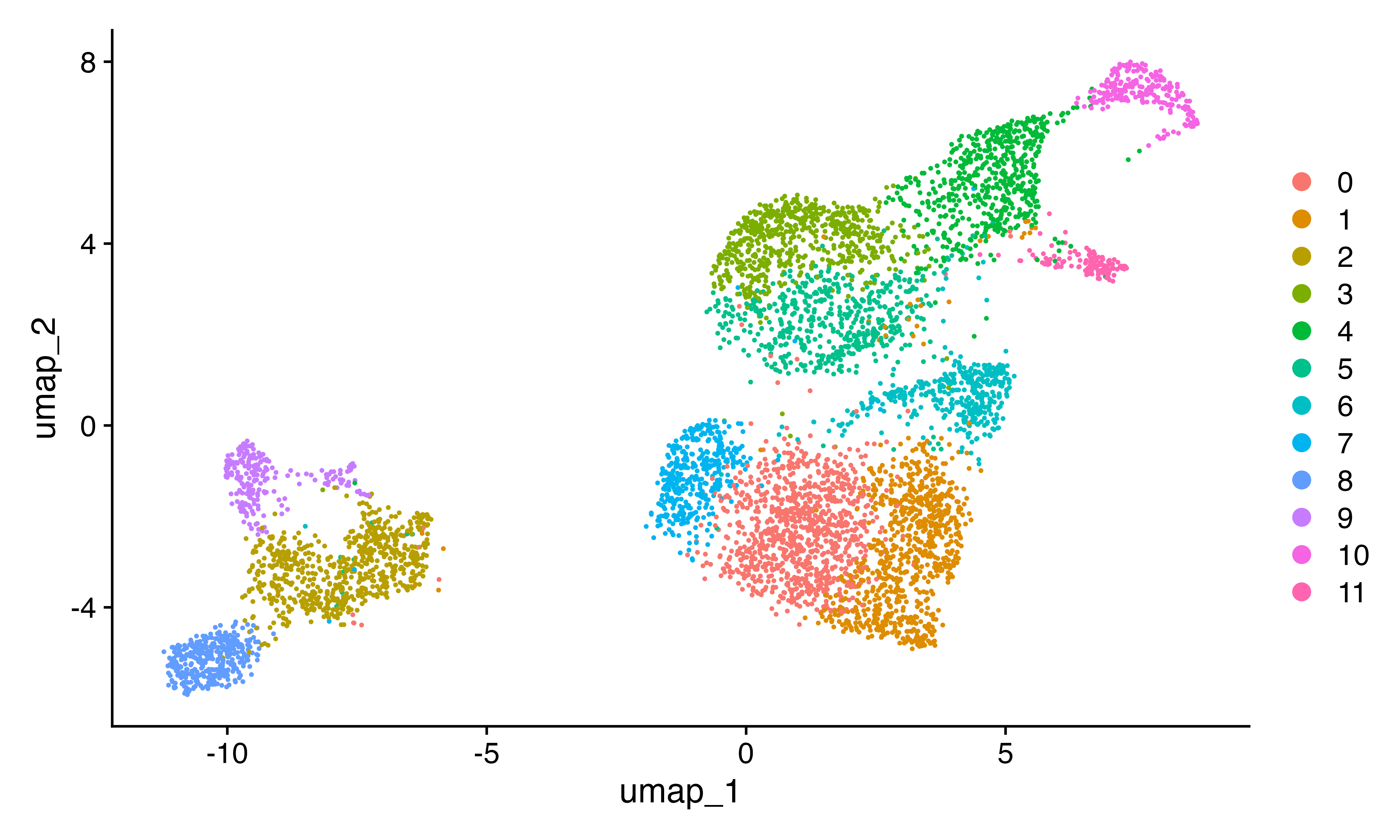
FeaturePlot_scCustom(seu, features = "CD4")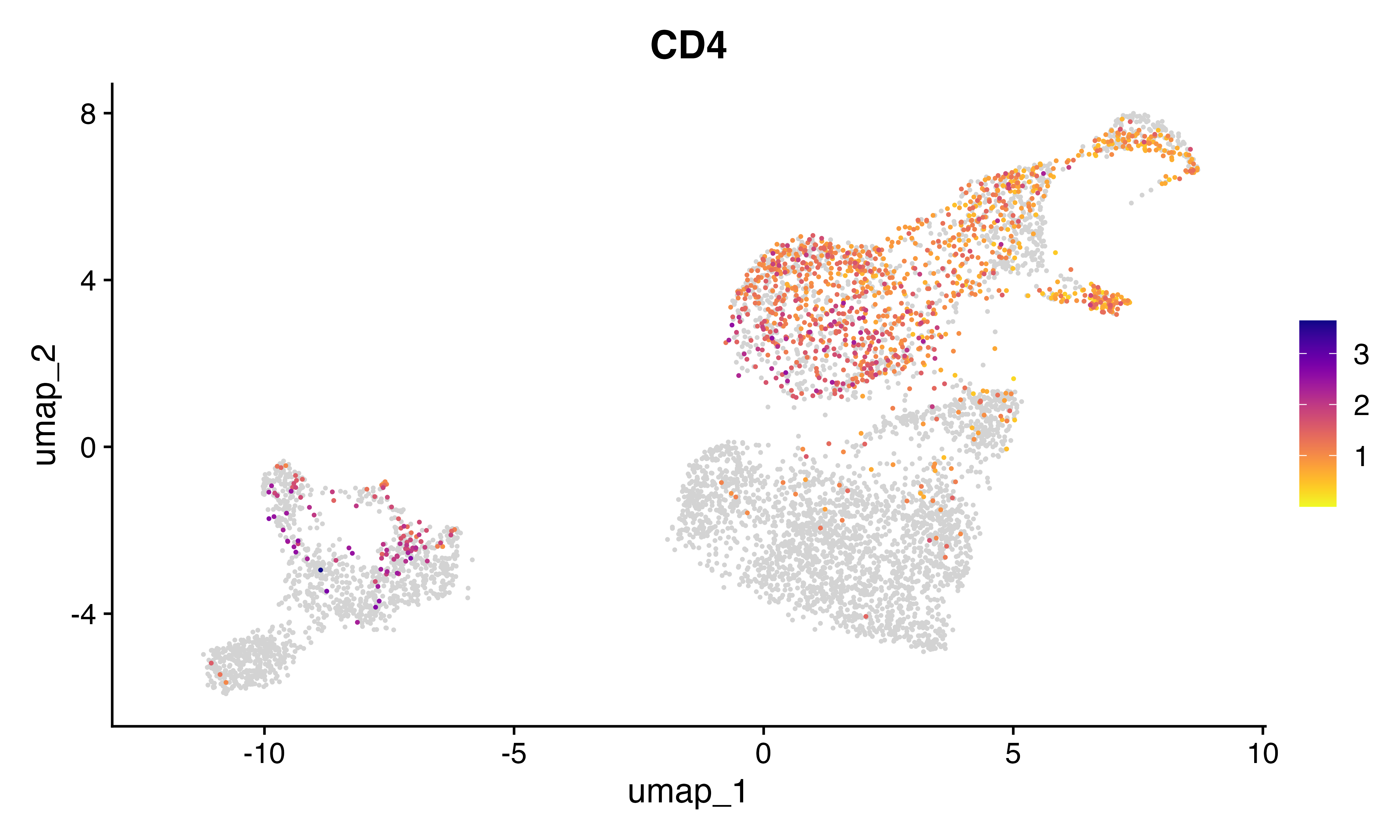
FeaturePlot_scCustom(seu, features = "MKI67")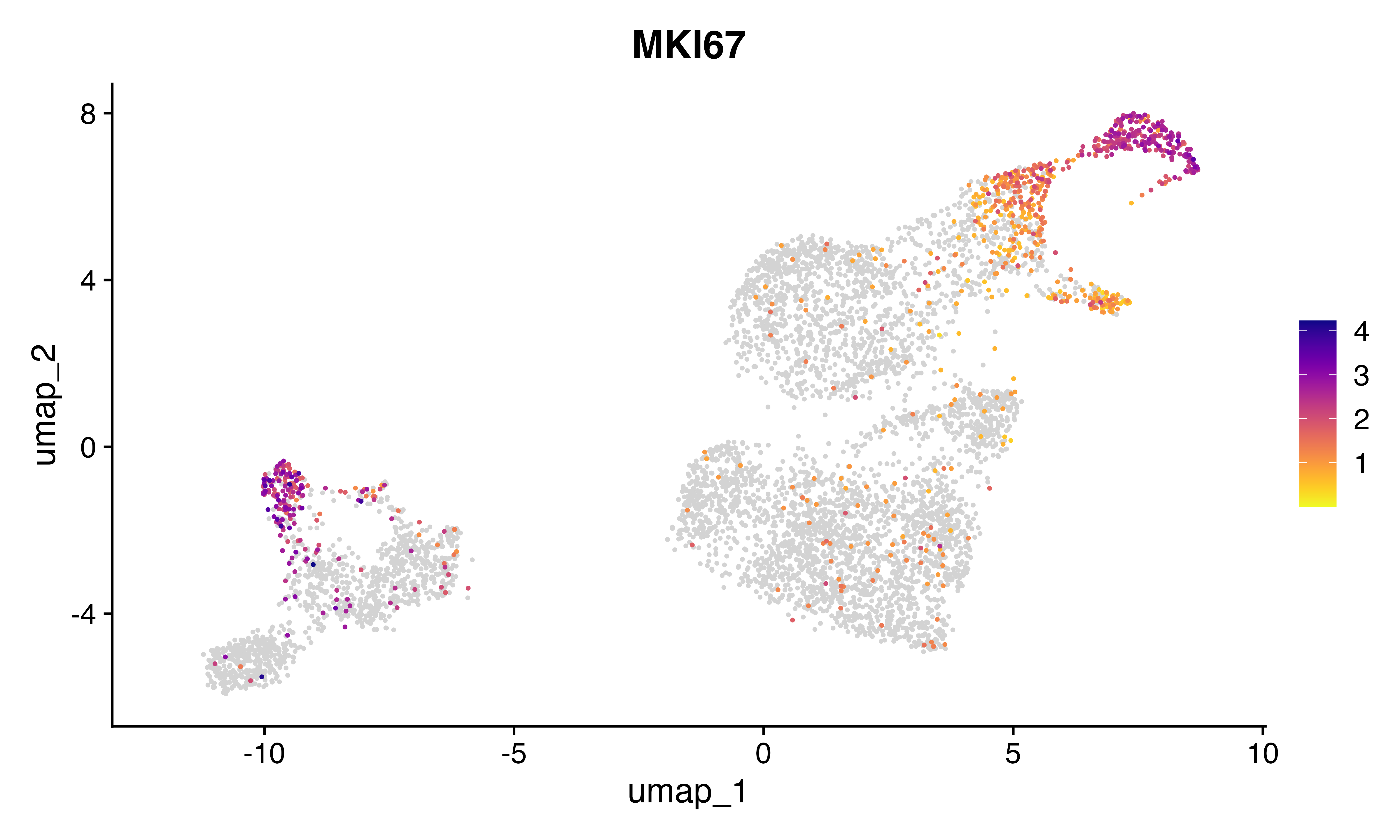
FeaturePlot_scCustom(seu, features = "CD8A")
FeaturePlot_scCustom(seu, features = "CAR")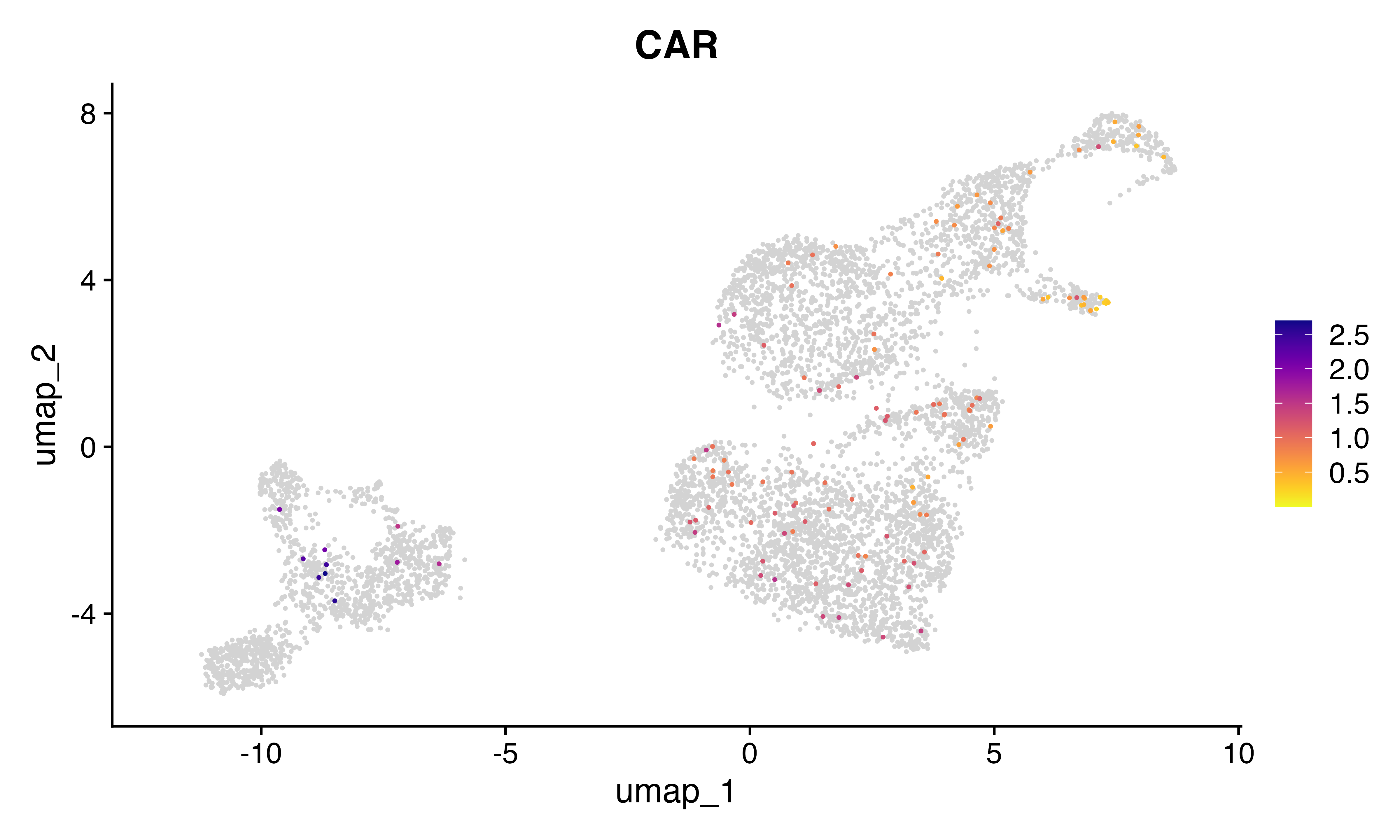
FeaturePlot_scCustom(seu, features = "FOXP3")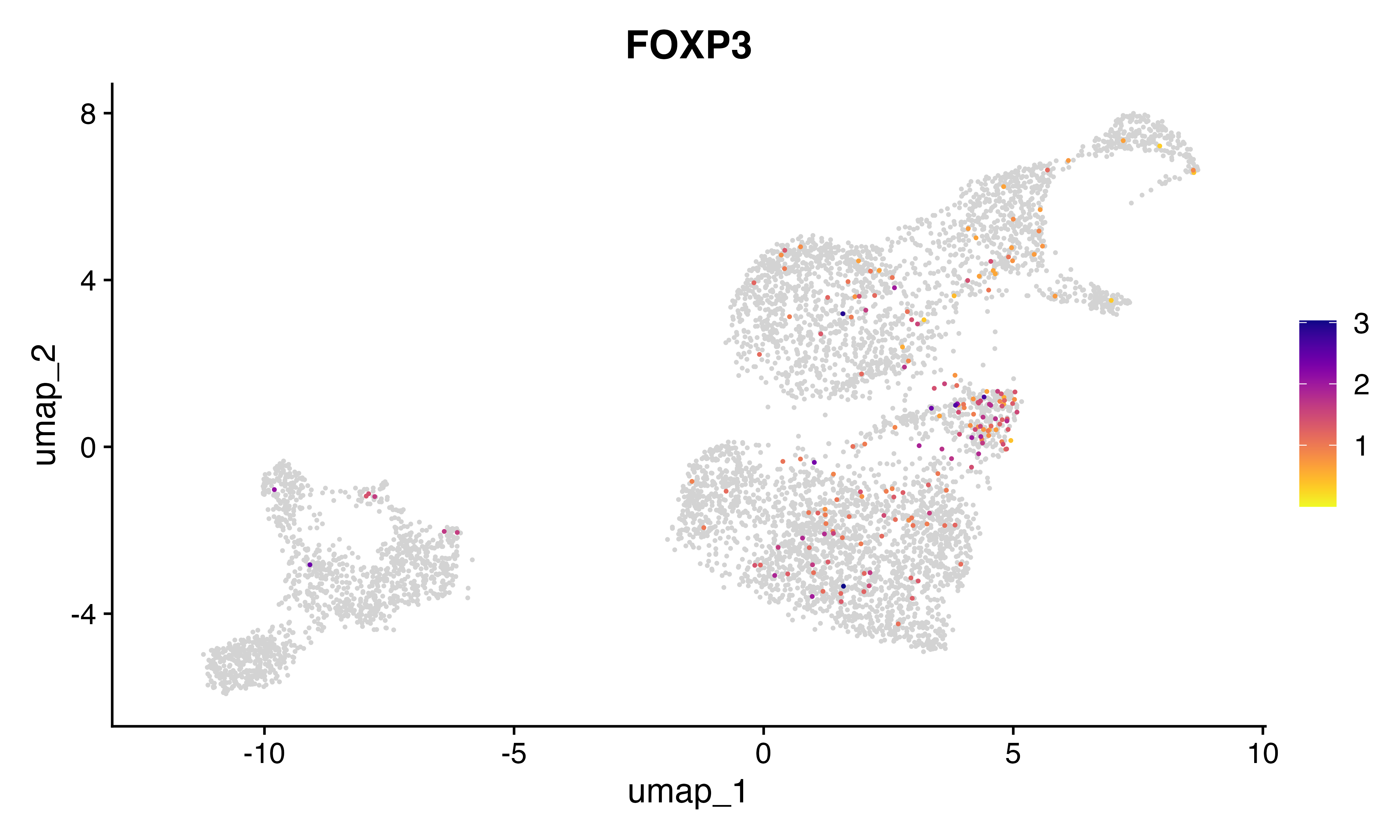
DimPlot_scCustom(seu, group.by = "geno_from2")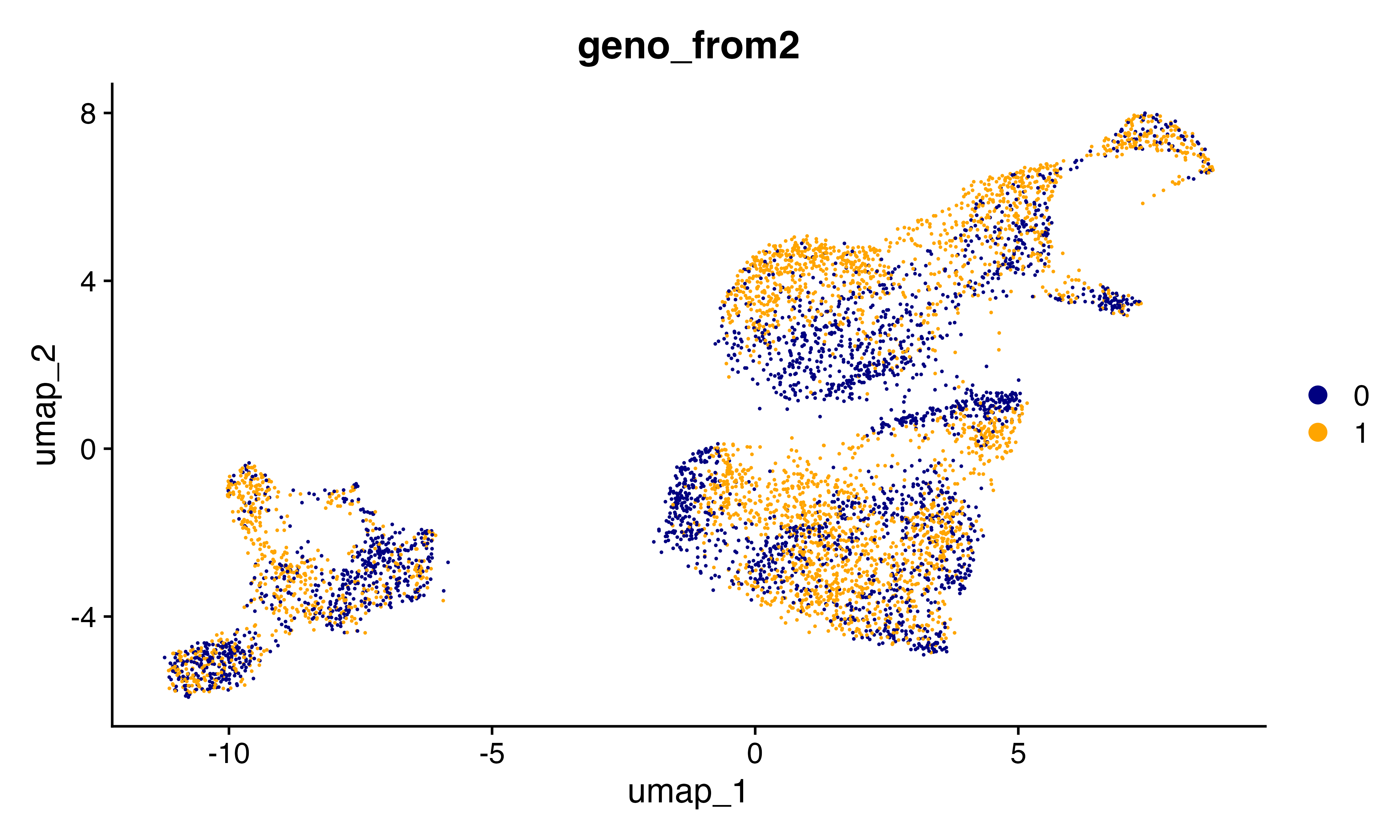
m<-FindAllMarkers(seu)
m[m$cluster==2,]## p_val avg_log2FC pct.1 pct.2 p_val_adj cluster
## RPS4X.1 6.645386e-273 -2.7620249 0.432 0.921 2.432344e-268 2
## RPL10.1 3.305150e-266 -2.7042673 0.683 0.949 1.209751e-261 2
## RPLP1.2 3.709151e-266 -2.5126705 0.668 0.950 1.357624e-261 2
## RPS14.2 1.251237e-263 -2.6522014 0.444 0.924 4.579777e-259 2
## RPS7.1 8.836598e-262 -2.6308882 0.371 0.917 3.234372e-257 2
## RPL41.2 5.026464e-261 -2.4504483 0.608 0.933 1.839786e-256 2
## RPL26.2 3.382796e-260 -2.5366981 0.414 0.924 1.238171e-255 2
## RPS13.2 2.215073e-258 -2.6041284 0.405 0.918 8.107612e-254 2
## RPS3A.1 2.302996e-257 -2.4480957 0.508 0.927 8.429427e-253 2
## RPL18A.1 4.334130e-257 -2.3191083 0.567 0.938 1.586378e-252 2
## RPS3.1 9.077122e-257 -2.3142906 0.580 0.935 3.322408e-252 2
## RPL30.2 9.438836e-257 -2.5247082 0.463 0.919 3.454803e-252 2
## RPL18.1 3.321796e-256 -2.3821516 0.460 0.926 1.215844e-251 2
## EEF1A1.1 1.397746e-253 -2.2486231 0.815 0.956 5.116028e-249 2
## RPS15A.2 1.429069e-252 -2.3783142 0.477 0.927 5.230678e-248 2
## RPS12.1 3.331363e-252 -2.3558259 0.653 0.945 1.219346e-247 2
## TMSB4X.2 6.400871e-252 -2.4434438 0.750 0.951 2.342847e-247 2
## RPS24.2 2.380323e-250 -2.4941160 0.371 0.912 8.712457e-246 2
## RPL19.1 2.744855e-250 -2.2397317 0.567 0.931 1.004672e-245 2
## RPL7A.1 4.453737e-250 -2.3143152 0.480 0.927 1.630157e-245 2
## RPL11.1 6.726701e-250 -2.2696574 0.544 0.929 2.462107e-245 2
## B2M.2 2.821742e-246 -2.0684415 0.859 0.961 1.032814e-241 2
## RPL14.1 7.675795e-245 -2.2800155 0.439 0.920 2.809495e-240 2
## RPS27A.2 7.072842e-244 -2.2829524 0.498 0.926 2.588802e-239 2
## RPL32.1 4.528932e-243 -2.2112383 0.550 0.931 1.657680e-238 2
## RPL15.1 2.697557e-242 -2.4535237 0.331 0.913 9.873596e-238 2
## RPL29.1 2.808600e-242 -2.2009646 0.501 0.925 1.028004e-237 2
## RPS5.2 4.364146e-242 -2.3952328 0.388 0.919 1.597365e-237 2
## RPL3.1 1.286902e-240 -2.1751942 0.547 0.926 4.710317e-236 2
## RPL8.1 1.435674e-240 -2.1860175 0.480 0.920 5.254855e-236 2
## FAU.2 4.027158e-239 -2.2972663 0.364 0.913 1.474021e-234 2
## RPL39.1 5.129921e-237 -2.5353541 0.289 0.904 1.877654e-232 2
## RPS28.1 1.522132e-236 -2.2569623 0.402 0.918 5.571309e-232 2
## RPL6.2 1.764600e-236 -2.3118296 0.391 0.915 6.458789e-232 2
## RPSA.2 6.464448e-236 -2.2758491 0.414 0.914 2.366117e-231 2
## RACK1.1 1.526521e-233 -2.2764629 0.391 0.909 5.587373e-229 2
## MT-CO1.1 1.395280e-232 0.6805214 0.998 0.999 5.107005e-228 2
## RPL9.2 1.745117e-232 -2.5712930 0.269 0.899 6.387479e-228 2
## RPS6.2 2.729877e-232 -2.2135675 0.475 0.922 9.991896e-228 2
## RPL5.1 3.346172e-231 -2.5278360 0.298 0.900 1.224766e-226 2
## RPL10A.1 5.415905e-231 -2.4310692 0.313 0.904 1.982330e-226 2
## RPS23.1 6.392986e-231 -2.1413859 0.513 0.926 2.339961e-226 2
## RPL34.1 6.687250e-231 -2.3217537 0.382 0.912 2.447667e-226 2
## RPS25.1 5.729251e-230 -2.2723575 0.365 0.911 2.097021e-225 2
## BTF3.1 7.718393e-228 -2.6410224 0.220 0.888 2.825086e-223 2
## MALAT1.1 1.276134e-226 1.3970551 0.998 0.943 4.670907e-222 2
## RPS2.2 2.176788e-223 -1.9702023 0.574 0.934 7.967480e-219 2
## RPS16.1 3.236686e-223 -2.1620780 0.346 0.919 1.184692e-218 2
## EEF1B2.2 7.214985e-222 -2.4140909 0.280 0.902 2.640829e-217 2
## RPLP0.2 1.534027e-221 -1.8859509 0.583 0.946 5.614847e-217 2
## RPL23A.1 5.042004e-220 -2.1720214 0.328 0.912 1.845474e-215 2
## NACA.2 1.442070e-219 -2.2691909 0.311 0.903 5.278264e-215 2
## RPS8.2 1.586115e-218 -1.9483371 0.623 0.932 5.805497e-214 2
## RPL35A.1 1.315926e-216 -2.2733390 0.283 0.903 4.816553e-212 2
## RPS9.1 1.216877e-214 -1.9203904 0.462 0.922 4.454012e-210 2
## SH3BGRL3.2 1.938926e-213 -2.1791101 0.405 0.909 7.096858e-209 2
## PFN1.1 2.322727e-213 -1.8925405 0.553 0.925 8.501646e-209 2
## RPS18.2 1.031575e-211 -1.8692774 0.662 0.945 3.775770e-207 2
## UBA52.2 7.349847e-211 -2.2708222 0.253 0.893 2.690191e-206 2
## RPL24.1 1.419595e-210 -2.1703810 0.287 0.904 5.196002e-206 2
## RPL28.1 2.304717e-210 -1.7537977 0.632 0.939 8.435725e-206 2
## MT-CO2.2 2.408172e-210 0.4909213 1.000 0.999 8.814390e-206 2
## RPL21.2 2.851301e-210 -2.2097728 0.287 0.901 1.043633e-205 2
## PPIA.1 1.052448e-208 -2.2287678 0.313 0.901 3.852169e-204 2
## IFITM1.1 1.430938e-208 -2.5998662 0.217 0.878 5.237521e-204 2
## COX4I1.2 2.034981e-207 -2.2989684 0.226 0.893 7.448439e-203 2
## GAPDH.1 1.533764e-206 -2.0771557 0.496 0.934 5.613882e-202 2
## TPT1.2 5.640029e-206 -1.6490306 0.713 0.948 2.064364e-201 2
## FTL.2 7.701798e-206 -2.1400715 0.394 0.909 2.819012e-201 2
## RPL12.2 1.163074e-205 -1.9484109 0.481 0.921 4.257083e-201 2
## RPLP2.1 3.669327e-205 -1.9196106 0.388 0.920 1.343047e-200 2
## RPL17.1 2.381208e-204 -1.9226557 0.394 0.914 8.715697e-200 2
## MT-CYB.2 1.159361e-202 0.7389710 1.000 0.998 4.243493e-198 2
## MYL12A.2 1.291825e-202 -2.1960194 0.305 0.894 4.728337e-198 2
## RPL22.1 1.072425e-201 -2.4017425 0.189 0.878 3.925289e-197 2
## RPL7.2 2.349907e-201 -2.4857570 0.179 0.868 8.601129e-197 2
## HINT1.2 1.878883e-200 -2.3195008 0.214 0.880 6.877087e-196 2
## ATP5MC2.2 7.219433e-200 -2.4768489 0.149 0.858 2.642457e-195 2
## CLIC1.2 2.612744e-197 -2.4433969 0.197 0.866 9.563165e-193 2
## EEF1G.2 9.186412e-196 -2.2173310 0.269 0.886 3.362410e-191 2
## MT-CO3.2 1.083242e-194 0.3637795 1.000 0.999 3.964882e-190 2
## RPL27.1 2.695058e-194 -2.2420328 0.202 0.879 9.864450e-190 2
## MT-ATP6.2 2.737100e-193 0.3280993 1.000 0.996 1.001833e-188 2
## RPL13.1 1.250977e-191 -1.5693698 0.765 0.958 4.578826e-187 2
## MYL6.1 2.023946e-190 -2.0791883 0.299 0.894 7.408047e-186 2
## ACTB.1 1.265869e-189 -1.7671473 0.811 0.956 4.633334e-185 2
## LDHB.2 6.016295e-189 -2.1947894 0.248 0.871 2.202084e-184 2
## RPL37.1 3.576396e-187 -1.8150955 0.367 0.908 1.309033e-182 2
## CD52.2 8.738011e-184 -2.1350431 0.296 0.888 3.198287e-179 2
## EIF3K.2 9.254536e-184 -2.2195446 0.167 0.852 3.387345e-179 2
## PFDN5.2 9.497484e-184 -2.0897487 0.180 0.876 3.476269e-179 2
## ATP5MG.2 3.053866e-183 -2.1715774 0.161 0.857 1.117776e-178 2
## ARPC3.2 3.267675e-183 -2.2298057 0.174 0.854 1.196035e-178 2
## IFITM2.2 7.264616e-183 -2.3502613 0.203 0.854 2.658995e-178 2
## CFL1.1 1.541872e-180 -1.7897452 0.426 0.904 5.643559e-176 2
## COX7C.2 2.147091e-180 -2.4049862 0.117 0.815 7.858783e-176 2
## RPS15.1 2.246499e-179 -1.5387063 0.541 0.927 8.222635e-175 2
## RPS27.2 2.271822e-179 -1.7024569 0.435 0.916 8.315322e-175 2
## RPS21.1 2.306682e-179 -1.9421048 0.250 0.892 8.442918e-175 2
## NDUFA4.1 1.304411e-177 -2.7750014 0.081 0.770 4.774404e-173 2
## RPL36AL.2 3.363108e-176 -2.1615344 0.177 0.860 1.230965e-171 2
## PSME1.2 2.487005e-175 -2.3660285 0.131 0.814 9.102937e-171 2
## MYL12B.2 6.328378e-175 -2.1579680 0.161 0.845 2.316313e-170 2
## RPS19.2 5.276602e-174 -1.4527535 0.743 0.953 1.931342e-169 2
## S100A4.2 3.569831e-173 -2.4830412 0.257 0.840 1.306630e-168 2
## RPS26.2 3.772058e-173 -2.0715530 0.247 0.876 1.380649e-168 2
## TMSB10.1 1.928844e-171 -1.5501133 0.603 0.937 7.059954e-167 2
## OST4.2 2.857021e-171 -2.5989191 0.069 0.750 1.045727e-166 2
## SNHG29.2 2.295151e-170 -2.2848535 0.125 0.809 8.400712e-166 2
## CHCHD2 2.502578e-169 -2.0297916 0.188 0.851 9.159937e-165 2
## NME2.1 3.038364e-168 -2.3056959 0.125 0.801 1.112102e-163 2
## RPS4Y1.2 1.251480e-167 -1.9726177 0.229 0.864 4.580667e-163 2
## ACTG1.1 3.220412e-167 -1.7188174 0.543 0.916 1.178735e-162 2
## NDUFB11.2 5.856568e-167 -2.7306856 0.050 0.709 2.143621e-162 2
## ATP5F1E.2 6.144481e-167 -1.9369867 0.141 0.838 2.249003e-162 2
## RNASEK.2 4.862057e-166 -2.1815158 0.108 0.792 1.779610e-161 2
## SSR4.2 5.010012e-166 -2.1073194 0.143 0.827 1.833765e-161 2
## COMMD6.2 5.885738e-166 -2.4969517 0.075 0.749 2.154298e-161 2
## S100A10.2 1.124133e-165 -2.0172794 0.155 0.831 4.114551e-161 2
## FTH1.1 1.194430e-165 -1.7566556 0.340 0.897 4.371853e-161 2
## SERF2.2 9.771259e-165 -1.6724884 0.308 0.906 3.576476e-160 2
## RPS17.1 2.166643e-164 -2.5707838 0.069 0.731 7.930345e-160 2
## IL32.2 2.557771e-164 -1.8007793 0.711 0.922 9.361955e-160 2
## TOMM7.2 5.707127e-164 -2.1759675 0.102 0.775 2.088923e-159 2
## CD3D.2 3.068605e-163 -1.9974613 0.165 0.834 1.123171e-158 2
## RPL35.1 5.845730e-163 -1.6371132 0.322 0.905 2.139654e-158 2
## HLA-B.2 1.056542e-162 -1.2497891 0.735 0.928 3.867153e-158 2
## SRP14.2 8.228054e-162 -2.0582197 0.149 0.825 3.011632e-157 2
## PPIB.2 4.086308e-161 -1.8915123 0.250 0.878 1.495670e-156 2
## S100A6.2 4.595512e-161 -1.8474516 0.259 0.881 1.682049e-156 2
## ATP5MC3.1 7.562286e-161 -2.4336540 0.110 0.768 2.767948e-156 2
## OAZ1.2 1.352707e-160 -1.9387186 0.220 0.857 4.951176e-156 2
## COX6A1.1 3.211757e-160 -2.1366188 0.104 0.778 1.175567e-155 2
## FXYD5.1 4.629815e-160 -1.9316387 0.253 0.866 1.694605e-155 2
## EMP3.2 8.525297e-160 -2.0231158 0.195 0.850 3.120429e-155 2
## GABARAP.2 1.437507e-157 -2.2350804 0.122 0.780 5.261563e-153 2
## SLC25A5 3.072680e-156 -2.4627856 0.093 0.732 1.124662e-151 2
## HCST.2 3.134846e-156 -2.2209557 0.131 0.777 1.147416e-151 2
## RPS29.1 3.275827e-156 -2.1622223 0.111 0.773 1.199018e-151 2
## CD3E.2 9.403247e-156 -1.7930867 0.263 0.879 3.441777e-151 2
## ATP5PO.2 6.064934e-155 -2.4303933 0.059 0.696 2.219887e-150 2
## HLA-A.2 1.581640e-154 -1.3517217 0.624 0.920 5.789118e-150 2
## COX6B1.2 2.552721e-154 -2.1234956 0.089 0.751 9.343469e-150 2
## RPL4.2 3.964475e-154 -1.6502178 0.335 0.889 1.451077e-149 2
## S100A11.2 6.728881e-154 -1.8408815 0.200 0.839 2.462905e-149 2
## RPL36.1 1.212905e-153 -1.5403605 0.325 0.904 4.439474e-149 2
## SEC61B.2 1.837950e-153 -2.2491348 0.104 0.759 6.727266e-149 2
## COX6C.1 3.920262e-153 -2.1823925 0.081 0.726 1.434894e-148 2
## RPL36A.1 1.062217e-152 -1.7634946 0.226 0.863 3.887928e-148 2
## HLA-C.2 1.717902e-151 -1.3238900 0.562 0.916 6.287865e-147 2
## RPL37A.1 1.832531e-150 -1.6826136 0.208 0.854 6.707430e-146 2
## TRAPPC1.2 6.899998e-150 -2.4738073 0.060 0.686 2.525537e-145 2
## COX8A.1 1.983150e-148 -2.1337021 0.105 0.748 7.258726e-144 2
## NDUFA11.1 6.944737e-148 -2.2803243 0.071 0.703 2.541913e-143 2
## SUMO2.1 7.626369e-148 -1.9927229 0.138 0.790 2.791403e-143 2
## HNRNPA1.2 1.721763e-147 -1.6272697 0.296 0.878 6.301998e-143 2
## H3F3A.2 1.783993e-147 -1.7308199 0.257 0.863 6.529770e-143 2
## PARK7.2 1.962013e-147 -2.4777823 0.080 0.709 7.181359e-143 2
## SNRPD2.2 5.808883e-147 -1.7476931 0.140 0.806 2.126167e-142 2
## RPL27A.1 6.358098e-146 -1.6936783 0.218 0.859 2.327191e-141 2
## PPDPF.2 8.992122e-146 -1.6116455 0.251 0.877 3.291297e-141 2
## RPL31.1 1.265978e-145 -1.5718220 0.223 0.863 4.633732e-141 2
## BRK1.2 2.626345e-145 -2.3989135 0.077 0.698 9.612947e-141 2
## LY6E.1 1.179174e-144 -2.0858081 0.144 0.777 4.316014e-140 2
## SLC25A6.2 1.370152e-144 -1.6878188 0.233 0.857 5.015030e-140 2
## RPS11.1 1.992094e-144 -1.6393254 0.235 0.865 7.291463e-140 2
## UQCRH.1 3.519165e-144 -2.1993546 0.072 0.694 1.288085e-139 2
## HIGD2A.2 7.835063e-144 -2.1770005 0.101 0.731 2.867790e-139 2
## TMA7.2 3.265274e-143 -1.6714238 0.135 0.792 1.195156e-138 2
## RAC2.2 4.583876e-143 -1.6262494 0.307 0.874 1.677790e-138 2
## COX5A 5.217225e-143 -2.1925105 0.086 0.709 1.909609e-138 2
## NEDD8.2 9.182346e-141 -2.5203294 0.066 0.673 3.360922e-136 2
## PTMA.1 2.750578e-140 -1.4567909 0.531 0.924 1.006767e-135 2
## CUTA.2 5.136810e-140 -2.3434637 0.060 0.669 1.880175e-135 2
## UBB.2 7.876840e-140 -1.6791703 0.140 0.785 2.883081e-135 2
## UQCRB.2 2.087879e-139 -2.0605592 0.077 0.695 7.642056e-135 2
## ABRACL.2 3.389116e-139 -1.7706527 0.110 0.741 1.240484e-134 2
## PRDX2 1.587457e-138 -2.3995292 0.081 0.685 5.810409e-134 2
## PPP1CA.2 6.700791e-138 -1.9439794 0.141 0.768 2.452624e-133 2
## RPL23.1 9.523175e-138 -1.7038982 0.134 0.769 3.485673e-133 2
## SSR2.2 1.304796e-137 -1.6287623 0.177 0.813 4.775813e-133 2
## CSTB.1 5.252172e-137 -1.8361890 0.110 0.730 1.922400e-132 2
## APRT.2 8.035756e-137 -1.7755586 0.131 0.762 2.941247e-132 2
## PSMB9.1 1.618490e-136 -1.8289760 0.143 0.768 5.923998e-132 2
## COX7A2.2 2.218008e-135 -2.1457207 0.075 0.675 8.118353e-131 2
## UQCR11.2 4.705109e-135 -1.9197994 0.081 0.692 1.722164e-130 2
## PCBP1.2 9.044602e-135 -1.9735115 0.116 0.737 3.310505e-130 2
## GMFG.2 2.225708e-134 -1.3538510 0.194 0.841 8.146537e-130 2
## EIF3F.2 1.499337e-133 -1.5463534 0.188 0.818 5.487872e-129 2
## UXT.1 4.301993e-133 -2.1269868 0.057 0.651 1.574616e-128 2
## MIF 4.783145e-133 -1.4576998 0.245 0.861 1.750727e-128 2
## PLAAT4.1 4.871701e-132 -1.7756478 0.165 0.776 1.783140e-127 2
## NDUFB9.1 3.406180e-131 -2.2130703 0.062 0.647 1.246730e-126 2
## C9orf16.2 4.920734e-131 -1.5504428 0.122 0.751 1.801087e-126 2
## MT-ND4L.2 7.799643e-131 0.2638235 0.953 0.969 2.854825e-126 2
## PSMB1.2 7.847405e-131 -2.1444325 0.084 0.677 2.872307e-126 2
## TOMM6.1 4.251883e-130 -2.7505297 0.041 0.607 1.556274e-125 2
## EEF2.1 4.999815e-130 -1.4239000 0.424 0.895 1.830032e-125 2
## NPM1.1 8.138638e-130 -1.4415197 0.344 0.885 2.978904e-125 2
## PSMB8.2 8.165767e-130 -1.6642779 0.143 0.762 2.988834e-125 2
## PSMA7.2 6.297441e-129 -1.7547826 0.116 0.724 2.304989e-124 2
## EIF5A 1.076543e-128 -1.8054926 0.119 0.720 3.940363e-124 2
## UBL5.2 1.460965e-128 -1.9433410 0.071 0.657 5.347424e-124 2
## ATP5PD.2 3.426487e-128 -2.2927688 0.048 0.618 1.254163e-123 2
## TRMT112.2 4.184805e-128 -1.9030754 0.072 0.661 1.531722e-123 2
## MTRNR2L12.2 7.094251e-128 0.3091380 0.977 0.990 2.596638e-123 2
## MT-ND5.2 2.618285e-127 0.4126632 0.956 0.970 9.583448e-123 2
## CD3G.2 6.055500e-127 -1.6943237 0.119 0.724 2.216434e-122 2
## TPI1 7.405296e-127 -1.7698393 0.153 0.755 2.710487e-122 2
## EDF1.2 9.370403e-127 -1.5032756 0.140 0.763 3.429755e-122 2
## SOD1 1.493381e-126 -2.0396423 0.069 0.645 5.466074e-122 2
## TAGLN2.2 1.530284e-126 -1.7358324 0.177 0.778 5.601144e-122 2
## ATP5PF.1 9.685690e-126 -2.1021588 0.057 0.626 3.545156e-121 2
## CD7.2 1.186079e-125 -1.7646729 0.221 0.797 4.341286e-121 2
## VAMP8.2 1.647961e-125 -2.2625009 0.060 0.631 6.031868e-121 2
## RBM3 5.447024e-125 -1.6851010 0.141 0.740 1.993720e-120 2
## PCED1B-AS1.2 8.647428e-125 -1.9219975 0.126 0.723 3.165132e-120 2
## KRTCAP2.2 8.872219e-125 -1.7759270 0.095 0.689 3.247410e-120 2
## SF3B5.2 2.257377e-124 -2.1474381 0.053 0.615 8.262451e-120 2
## CDK2AP2.2 3.761757e-124 -1.8765923 0.110 0.698 1.376878e-119 2
## GYPC.2 3.841903e-124 -1.6430502 0.135 0.745 1.406213e-119 2
## SPCS1.2 6.374914e-124 -1.8294817 0.095 0.681 2.333346e-119 2
## RPS20 1.167015e-123 -1.5035075 0.110 0.715 4.271509e-119 2
## EIF3E.1 3.454257e-123 -1.7363329 0.099 0.688 1.264327e-118 2
## MT-ND1.2 1.495542e-122 0.4249429 0.943 0.967 5.473983e-118 2
## CRIP1.2 1.726592e-122 -1.7368262 0.144 0.740 6.319671e-118 2
## ATP6V0E1.2 3.760815e-122 -1.6607912 0.090 0.675 1.376533e-117 2
## ERH.2 6.417596e-122 -1.9514263 0.071 0.634 2.348969e-117 2
## SEC61G.2 8.334467e-122 -2.2457453 0.056 0.612 3.050582e-117 2
## PSMB3.1 1.111533e-121 -2.2593164 0.059 0.613 4.068435e-117 2
## NDUFB8.2 6.319919e-121 -1.6203586 0.113 0.710 2.313217e-116 2
## TLE5.2 9.194342e-121 -1.3658278 0.253 0.851 3.365313e-116 2
## ATP5PB.2 7.520043e-120 -1.6445088 0.099 0.681 2.752486e-115 2
## NDUFC2.1 1.710979e-119 -2.1700741 0.056 0.607 6.262526e-115 2
## RAN 4.029657e-119 -1.6496015 0.144 0.732 1.474935e-114 2
## GNG5.2 7.012997e-119 -1.9281139 0.068 0.624 2.566897e-114 2
## SUB1.2 9.817525e-119 -1.4663794 0.188 0.787 3.593410e-114 2
## ERP29.2 5.038650e-118 -1.6596457 0.114 0.700 1.844247e-113 2
## TIMP1.2 1.603307e-117 -2.0667655 0.105 0.667 5.868423e-113 2
## ARF5.2 3.616858e-117 -2.0646246 0.077 0.633 1.323842e-112 2
## EIF3L.1 8.908085e-117 -1.5947302 0.131 0.715 3.260537e-112 2
## ATP5MF.2 9.497577e-117 -2.2291333 0.051 0.593 3.476303e-112 2
## EIF3G.2 1.721758e-116 -1.5299663 0.138 0.731 6.301977e-112 2
## DBI.2 2.289327e-116 -2.0045679 0.056 0.599 8.379396e-112 2
## COX5B.1 4.679224e-116 -1.3297416 0.107 0.700 1.712690e-111 2
## RHOA.1 5.585584e-116 -1.3405654 0.211 0.812 2.044435e-111 2
## GPX4 5.760640e-116 -1.7651610 0.086 0.649 2.108510e-111 2
## NDUFS5.2 7.255229e-116 -1.7930561 0.068 0.623 2.655559e-111 2
## RABAC1.2 1.536254e-115 -1.9436563 0.090 0.648 5.622998e-111 2
## FIS1.2 2.311379e-115 -2.0904568 0.057 0.597 8.460108e-111 2
## RPS27L.2 1.142076e-114 -1.9456466 0.101 0.662 4.180225e-110 2
## CSNK2B.2 1.521339e-114 -1.5226825 0.098 0.670 5.568406e-110 2
## DYNLL1.1 2.193558e-114 -1.4765938 0.117 0.691 8.028859e-110 2
## ARHGDIB.2 2.270775e-114 -1.2630196 0.402 0.891 8.311490e-110 2
## H2AFZ.1 2.411795e-114 -2.0204005 0.102 0.659 8.827650e-110 2
## WDR83OS.2 2.602713e-114 -1.9564498 0.063 0.610 9.526451e-110 2
## RSL1D1.1 9.286705e-114 -1.7792881 0.080 0.628 3.399120e-109 2
## CD48.2 1.420480e-113 -1.5134449 0.158 0.744 5.199242e-109 2
## PSME2.1 2.321075e-113 -1.5221339 0.167 0.748 8.495599e-109 2
## NDUFA12.2 2.616764e-113 -2.4374080 0.056 0.584 9.577879e-109 2
## ARPC1B.2 1.246459e-112 -1.2686426 0.289 0.853 4.562291e-108 2
## UQCR10.1 1.249133e-112 -1.9448571 0.069 0.610 4.572076e-108 2
## GUK1.2 2.010630e-112 -1.1502642 0.144 0.740 7.359308e-108 2
## CD99.2 1.222615e-111 -1.4712912 0.197 0.784 4.475015e-107 2
## TALDO1.1 3.103521e-111 -1.8852973 0.078 0.620 1.135951e-106 2
## RPL38.1 4.894694e-111 -1.2287225 0.147 0.745 1.791556e-106 2
## OSTC.2 5.655232e-111 -2.1894488 0.065 0.592 2.069928e-106 2
## NDUFB2.1 2.329009e-110 -1.9754641 0.063 0.596 8.524639e-106 2
## TRIR.2 2.699133e-110 -1.3687980 0.174 0.766 9.879367e-106 2
## TXN 4.871096e-110 -1.5289253 0.098 0.650 1.782918e-105 2
## PSMA2.1 2.458466e-109 -2.1439904 0.065 0.590 8.998477e-105 2
## DAD1.1 6.593497e-109 -1.4542356 0.131 0.705 2.413352e-104 2
## YBX1.1 1.852598e-108 -1.4584524 0.188 0.763 6.780878e-104 2
## BST2.1 2.698523e-108 -1.7445118 0.080 0.617 9.877133e-104 2
## PRR13.2 3.739548e-108 -1.6692318 0.092 0.636 1.368749e-103 2
## NDUFA13.1 4.462562e-108 -1.1525544 0.165 0.766 1.633387e-103 2
## CYBA.2 5.106788e-108 -1.2542201 0.367 0.883 1.869187e-103 2
## BAX.2 5.666374e-108 -1.4536091 0.182 0.755 2.074006e-103 2
## C19orf53.2 6.806067e-108 -1.5559509 0.078 0.619 2.491157e-103 2
## ATP5IF1.1 1.233507e-107 -1.6088133 0.084 0.627 4.514884e-103 2
## PRDX5.1 2.256476e-107 -1.6868417 0.096 0.637 8.259153e-103 2
## MZT2B.1 3.873875e-107 -1.3998067 0.129 0.700 1.417916e-102 2
## NOP10.1 4.138139e-107 -2.0786541 0.054 0.568 1.514642e-102 2
## GPSM3.2 5.112626e-107 -1.2006757 0.197 0.790 1.871323e-102 2
## PSMB6.1 7.769409e-107 -1.9771311 0.059 0.576 2.843759e-102 2
## RPL13A.2 1.736697e-106 -1.0844577 0.442 0.905 6.356657e-102 2
## ATP5F1B.2 2.241807e-106 -1.4207051 0.182 0.757 8.205463e-102 2
## MDH2.2 3.653114e-106 -1.7026840 0.081 0.613 1.337113e-101 2
## ENO1.2 4.483708e-106 -1.4111963 0.238 0.796 1.641127e-101 2
## ATP5F1D.2 1.954967e-105 -1.2337907 0.164 0.745 7.155571e-101 2
## C4orf3.2 2.250871e-105 -2.0996333 0.056 0.565 8.238638e-101 2
## YWHAB.2 3.858360e-105 -1.2996289 0.195 0.776 1.412237e-100 2
## CHCHD10.2 5.450032e-105 -1.7617163 0.080 0.606 1.994821e-100 2
## PHB2.1 8.059261e-105 -1.7978054 0.074 0.594 2.949851e-100 2
## PRELID1.1 9.237024e-105 -1.5501637 0.110 0.655 3.380935e-100 2
## DAZAP2.2 1.278890e-104 -1.3500446 0.147 0.721 4.680994e-100 2
## LSM7.1 1.321142e-104 -1.7859762 0.066 0.586 4.835643e-100 2
## GNB2.2 3.737274e-104 -1.5087991 0.101 0.640 1.367917e-99 2
## HMGN2.1 4.291114e-104 -1.8351194 0.141 0.688 1.570634e-99 2
## MT-ND4.2 4.868028e-104 0.3831482 0.935 0.969 1.781796e-99 2
## BANF1 7.448985e-104 -2.1851024 0.038 0.529 2.726478e-99 2
## SELENOH 1.282093e-103 -1.3727703 0.111 0.659 4.692718e-99 2
## C1QBP.1 3.221137e-103 -2.1016110 0.047 0.543 1.179001e-98 2
## NDUFA1.1 3.815533e-103 -1.7667708 0.060 0.571 1.396562e-98 2
## TAF10.2 1.334865e-102 -1.4152190 0.123 0.675 4.885875e-98 2
## SRP9.1 3.357929e-102 -1.3932323 0.093 0.623 1.229069e-97 2
## CD63.2 1.579859e-101 -2.0974691 0.083 0.592 5.782600e-97 2
## UBE2L6.1 1.667725e-101 -2.1427869 0.051 0.543 6.104208e-97 2
## ANAPC11 4.708647e-101 -2.6733530 0.035 0.512 1.723459e-96 2
## ELOB.1 5.851641e-101 -1.1556376 0.110 0.656 2.141818e-96 2
## ATP6V1G1.2 6.013061e-101 -1.3422621 0.137 0.690 2.200901e-96 2
## RTRAF.2 6.307559e-101 -1.4635145 0.119 0.659 2.308693e-96 2
## CIB1.2 7.119518e-101 -1.4345750 0.087 0.610 2.605886e-96 2
## EEF1D.2 1.250870e-100 -1.0714973 0.391 0.895 4.578433e-96 2
## LAMTOR4.2 2.441860e-100 -1.5209976 0.090 0.612 8.937697e-96 2
## GTF3A.1 3.615072e-100 -1.6356602 0.078 0.588 1.323189e-95 2
## TSPO.2 8.774382e-100 -2.1365899 0.050 0.538 3.211599e-95 2
## LTB.2 1.106812e-99 -1.2647145 0.657 0.910 4.051152e-95 2
## ATP5MD.1 2.274046e-99 -2.2565481 0.041 0.522 8.323462e-95 2
## DRAP1.2 2.605082e-99 -1.0683392 0.149 0.712 9.535121e-95 2
## MZT2A.2 3.779639e-99 -1.5688238 0.081 0.590 1.383424e-94 2
## SLC25A3.2 4.455643e-99 -1.3263166 0.188 0.743 1.630854e-94 2
## COX7A2L.2 4.730160e-99 -1.9653302 0.054 0.545 1.731333e-94 2
## SELENOW.1 1.686598e-98 -1.6087076 0.083 0.592 6.173285e-94 2
## ANP32B.1 2.337441e-98 -1.3062732 0.153 0.704 8.555502e-94 2
## GSTP1.1 3.527023e-98 -1.9201563 0.066 0.560 1.290961e-93 2
## LAPTM5.2 4.217307e-98 -1.2236159 0.271 0.828 1.543619e-93 2
## SNU13.2 4.960028e-98 -1.1210053 0.108 0.644 1.815469e-93 2
## GAS5.1 5.520780e-98 -1.1982491 0.180 0.739 2.020716e-93 2
## TSTD1.2 6.718713e-98 -2.8481569 0.020 0.476 2.459183e-93 2
## IL2RG.2 1.651513e-97 -1.1729696 0.283 0.843 6.044869e-93 2
## UBE2I.2 1.867841e-97 -1.4629135 0.072 0.573 6.836672e-93 2
## SMDT1.2 2.616029e-97 -1.9836260 0.050 0.531 9.575189e-93 2
## RGS10.2 2.974231e-97 -1.3241581 0.096 0.617 1.088628e-92 2
## COX7B.1 3.171545e-97 -1.8552750 0.059 0.547 1.160849e-92 2
## PSMB10.1 5.081488e-97 -1.2602776 0.119 0.650 1.859926e-92 2
## PSMA5.1 6.925576e-97 -1.2835046 0.107 0.637 2.534899e-92 2
## POMP.1 8.148264e-97 -1.6517980 0.065 0.559 2.982427e-92 2
## CCNI.2 1.617289e-96 -1.2126313 0.161 0.718 5.919599e-92 2
## ATP6V0B.1 1.809057e-96 -1.8309195 0.056 0.541 6.621511e-92 2
## SNRPB 1.848075e-96 -1.7605204 0.090 0.593 6.764323e-92 2
## PSMD8.1 1.962760e-96 -1.5769545 0.075 0.573 7.184095e-92 2
## C12orf57.2 2.734206e-96 -1.0771720 0.143 0.689 1.000774e-91 2
## NDUFB10.2 3.067811e-96 -1.3004753 0.068 0.565 1.122880e-91 2
## SAP18.2 3.743388e-96 -1.2836890 0.101 0.623 1.370155e-91 2
## MRPL51.1 4.507835e-96 -1.8814002 0.038 0.507 1.649958e-91 2
## ANAPC16.2 6.472762e-96 -1.2417109 0.092 0.604 2.369160e-91 2
## JTB.2 9.067079e-96 -1.5372120 0.078 0.579 3.318732e-91 2
## UCP2.2 3.627692e-95 -1.3257393 0.155 0.694 1.327808e-90 2
## PEBP1 4.437596e-95 -1.4245161 0.090 0.595 1.624249e-90 2
## CALM3.2 5.356481e-95 -1.4771017 0.104 0.615 1.960579e-90 2
## PRDX6.2 9.098964e-95 -1.6022162 0.084 0.585 3.330403e-90 2
## ATP6V0C.2 1.031789e-94 -1.6328248 0.075 0.570 3.776553e-90 2
## LGALS1.2 1.176406e-94 -1.6138253 0.221 0.719 4.305882e-90 2
## EIF3I 5.366624e-94 -1.5922898 0.069 0.556 1.964292e-89 2
## ATP6V1F.2 5.776862e-94 -2.1157467 0.044 0.508 2.114447e-89 2
## PGLS.2 6.485862e-94 -1.9171555 0.053 0.527 2.373955e-89 2
## DDOST.2 1.331595e-93 -1.5770678 0.116 0.628 4.873903e-89 2
## CNBP.2 1.470482e-93 -1.1401350 0.192 0.747 5.382259e-89 2
## TMEM258.2 1.757918e-93 -1.4333674 0.081 0.577 6.434331e-89 2
## LDHA.1 1.870631e-93 -1.4006553 0.158 0.684 6.846883e-89 2
## NDUFAB1.1 2.055473e-93 -1.8713191 0.045 0.512 7.523441e-89 2
## KRT10.2 3.081124e-93 -1.9472391 0.042 0.506 1.127753e-88 2
## ATP5F1A.2 3.399872e-93 -1.3331181 0.137 0.663 1.244421e-88 2
## PGK1 6.191160e-93 -1.2187410 0.185 0.730 2.266089e-88 2
## FBL.1 8.083614e-93 -1.7004603 0.069 0.551 2.958764e-88 2
## CD37.2 1.233694e-92 -1.1504213 0.235 0.791 4.515567e-88 2
## MAL.2 1.473979e-92 -1.9897196 0.071 0.546 5.395058e-88 2
## NOSIP.1 2.905230e-92 -1.1512101 0.119 0.637 1.063372e-87 2
## SF3B6.2 4.857363e-92 -1.7494105 0.045 0.507 1.777892e-87 2
## UBXN1.2 5.177986e-92 -1.0572946 0.126 0.659 1.895247e-87 2
## NDUFS7.2 1.141355e-91 -1.5865045 0.053 0.522 4.177589e-87 2
## SOS1.1 1.306783e-91 1.8600107 0.761 0.786 4.783086e-87 2
## TUBB.2 1.753833e-91 -2.0838534 0.128 0.628 6.419379e-87 2
## ICAM3.2 2.856339e-91 -1.1660291 0.155 0.695 1.045477e-86 2
## UFC1.2 5.660819e-91 -1.1285435 0.113 0.628 2.071973e-86 2
## TOMM22.2 6.456540e-91 -2.4319561 0.033 0.481 2.363223e-86 2
## CD27.2 6.991195e-91 -1.8643446 0.072 0.547 2.558917e-86 2
## PYCARD.2 7.422517e-91 -1.8332725 0.059 0.524 2.716790e-86 2
## PHPT1.1 8.381304e-91 -1.3693981 0.077 0.562 3.067725e-86 2
## CIAO2B.1 2.933894e-90 -1.2733825 0.081 0.570 1.073864e-85 2
## GABARAPL2.2 4.301402e-90 -1.7635516 0.059 0.526 1.574399e-85 2
## SNHG6.1 7.621346e-90 -1.3190549 0.060 0.529 2.789565e-85 2
## SNRPC 2.212597e-89 -2.4694200 0.029 0.468 8.098548e-85 2
## IFITM3 2.872414e-89 -2.6134371 0.036 0.476 1.051361e-84 2
## GIMAP4.2 6.465282e-89 -1.2702041 0.131 0.641 2.366422e-84 2
## ITM2B.2 8.103955e-89 -1.0368268 0.262 0.817 2.966210e-84 2
## RASAL2.1 1.970565e-88 -3.8068228 0.024 0.447 7.212662e-84 2
## FKBP1A.1 2.101404e-88 -1.7306217 0.077 0.550 7.691558e-84 2
## PGAM1.2 2.717062e-88 -1.5905212 0.092 0.570 9.944989e-84 2
## ARPC4.1 4.803246e-88 -1.1570744 0.152 0.678 1.758084e-83 2
## SIT1.1 4.891948e-88 -1.3632212 0.093 0.577 1.790551e-83 2
## SSBP1 4.922232e-88 -1.2669325 0.074 0.549 1.801635e-83 2
## DDT.1 8.993529e-88 -2.8251434 0.021 0.446 3.291812e-83 2
## POLR2L.2 1.175026e-87 -1.4297119 0.069 0.537 4.300829e-83 2
## RAC1.1 1.431400e-87 -1.4732398 0.095 0.580 5.239212e-83 2
## NDUFB4 2.410972e-87 -1.6271433 0.051 0.506 8.824641e-83 2
## NDUFA2 3.047454e-87 -1.6001697 0.044 0.488 1.115429e-82 2
## ERGIC3.2 7.092520e-87 -1.7521296 0.053 0.504 2.596004e-82 2
## COPE.2 7.680757e-87 -0.9522209 0.158 0.686 2.811311e-82 2
## OCIAD2.2 8.357415e-87 -1.9530500 0.053 0.500 3.058981e-82 2
## NHP2 1.336408e-86 -2.3533068 0.030 0.459 4.891519e-82 2
## EIF3H.1 1.965015e-86 -1.0535773 0.209 0.756 7.192346e-82 2
## GZMM.2 2.303708e-86 -1.4998699 0.095 0.569 8.432032e-82 2
## RBX1.1 2.979461e-86 -1.2417044 0.084 0.564 1.090542e-81 2
## ATP5F1C.1 2.991128e-86 -1.3077013 0.098 0.587 1.094813e-81 2
## CAPNS1.2 3.871743e-86 -1.4466567 0.086 0.559 1.417135e-81 2
## DCTN3.2 3.951792e-86 -2.2057101 0.036 0.470 1.446435e-81 2
## LAMTOR5.1 7.229984e-86 -1.6558142 0.057 0.508 2.646319e-81 2
## ARPC5.1 7.970888e-86 -1.1824196 0.185 0.716 2.917504e-81 2
## HSPA8.1 9.858774e-86 -1.0113885 0.460 0.899 3.608508e-81 2
## PPA1 1.258708e-85 -1.4320224 0.071 0.534 4.607123e-81 2
## SEC11C.2 2.310533e-85 -2.1592612 0.050 0.490 8.457014e-81 2
## MRPS21.2 4.299204e-85 -1.4418505 0.071 0.534 1.573595e-80 2
## TRBC2.2 5.161482e-85 -1.2522463 0.152 0.662 1.889206e-80 2
## MICOS10.2 8.232445e-85 -1.2618671 0.084 0.558 3.013239e-80 2
## C17orf49.2 1.166685e-84 -1.7268237 0.051 0.495 4.270302e-80 2
## CD81.2 1.346590e-84 -1.4684885 0.108 0.588 4.928787e-80 2
## ISCU.2 1.383907e-84 -2.0204827 0.038 0.469 5.065376e-80 2
## SNRPG.2 2.996416e-84 -1.4399924 0.071 0.530 1.096748e-79 2
## ALKBH7.2 3.849594e-84 -1.6594184 0.051 0.490 1.409028e-79 2
## TBCA.1 5.770097e-84 -1.0907152 0.120 0.617 2.111971e-79 2
## UQCRFS1.2 6.205186e-84 -1.4422656 0.072 0.533 2.271222e-79 2
## TUFM.2 7.147801e-84 -1.4580363 0.072 0.528 2.616238e-79 2
## NDUFA7.1 1.698179e-83 -1.9756957 0.039 0.468 6.215676e-79 2
## PSMA6.1 2.162362e-83 -1.2444865 0.095 0.569 7.914677e-79 2
## VAMP5.1 2.267251e-83 -1.7924536 0.053 0.492 8.298591e-79 2
## UQCRQ.1 2.394805e-83 -1.7347582 0.048 0.484 8.765467e-79 2
## TMEM50A.2 2.992116e-83 -1.0772936 0.134 0.640 1.095174e-78 2
## NSA2.1 3.133835e-83 -1.4152657 0.065 0.516 1.147046e-78 2
## CCT6A 4.211622e-83 -1.4207967 0.084 0.548 1.541538e-78 2
## PRDX1 8.830083e-83 -1.5065167 0.096 0.563 3.231987e-78 2
## PLP2.2 8.861350e-83 -1.5459101 0.068 0.517 3.243431e-78 2
## SRSF9.1 1.529639e-82 -1.2089319 0.125 0.618 5.598785e-78 2
## BCAP31.1 3.610209e-82 -1.3077333 0.090 0.556 1.321409e-77 2
## TWF2.2 4.665395e-82 -1.3200557 0.084 0.549 1.707628e-77 2
## NDUFS8.1 5.865026e-82 -1.3285576 0.075 0.530 2.146717e-77 2
## CORO1A.2 8.359623e-82 -0.9436258 0.498 0.904 3.059789e-77 2
## GNAI2.2 1.525566e-81 -0.9374541 0.214 0.744 5.583876e-77 2
## TXNL4A.2 1.528127e-81 -1.1130301 0.099 0.573 5.593249e-77 2
## SSR3.1 1.667010e-81 -1.3381873 0.105 0.578 6.101591e-77 2
## CNN2.2 2.127226e-81 -1.0057734 0.223 0.749 7.786073e-77 2
## SNX3.1 2.315182e-81 -1.5633987 0.063 0.505 8.474029e-77 2
## HSPE1 2.433525e-81 -1.3737694 0.089 0.547 8.907186e-77 2
## NDUFB3.1 2.659401e-81 -1.6662637 0.056 0.492 9.733941e-77 2
## SERP1.1 4.079148e-81 -0.9165754 0.211 0.747 1.493050e-76 2
## ARRB2.2 7.231477e-81 -1.0427509 0.131 0.617 2.646865e-76 2
## CALM1.2 1.053784e-80 -0.9171092 0.414 0.883 3.857062e-76 2
## TBCB.1 1.136480e-80 -1.3012176 0.084 0.540 4.159744e-76 2
## UBE2L3.1 1.448345e-80 -1.3427603 0.068 0.508 5.301232e-76 2
## NOP53.1 1.634923e-80 -0.8975550 0.227 0.765 5.984146e-76 2
## LMAN2.2 1.782289e-80 -1.2228470 0.101 0.569 6.523534e-76 2
## VPS28.2 2.695326e-80 -1.1018942 0.081 0.536 9.865431e-76 2
## PPP4C.1 2.766476e-80 -1.2486576 0.087 0.545 1.012585e-75 2
## GPI.2 3.941765e-80 -1.3330874 0.096 0.557 1.442765e-75 2
## CST7.2 4.877360e-80 -1.9310124 0.081 0.518 1.785211e-75 2
## SDHC.1 6.458222e-80 -1.5527735 0.059 0.493 2.363838e-75 2
## GHITM.1 7.294540e-80 -1.3337205 0.074 0.519 2.669948e-75 2
## HMGN1 3.129626e-79 -1.0524403 0.147 0.640 1.145506e-74 2
## ARL6IP4.2 4.056297e-79 -0.7098858 0.153 0.665 1.484686e-74 2
## TOMM20.2 4.756089e-79 -1.3100760 0.098 0.559 1.740824e-74 2
## ARPC2.2 5.717333e-79 -0.9437346 0.358 0.855 2.092658e-74 2
## MDH1.1 5.957987e-79 -1.6087717 0.077 0.520 2.180743e-74 2
## ST13 1.190240e-78 -1.3254609 0.077 0.520 4.356518e-74 2
## SF3B4.1 1.431495e-78 -2.2629376 0.033 0.438 5.239559e-74 2
## SEC11A.2 1.811796e-78 -1.1389114 0.089 0.544 6.631536e-74 2
## LSM2 2.278153e-78 -2.3949893 0.029 0.430 8.338496e-74 2
## SEM1.1 5.510431e-78 -1.0374992 0.080 0.527 2.016928e-73 2
## GZMA.1 5.669535e-78 -1.9548625 0.205 0.621 2.075163e-73 2
## RSL24D1.2 6.581152e-78 -1.5867153 0.056 0.478 2.408833e-73 2
## COTL1.2 8.538542e-78 -1.0225019 0.290 0.802 3.125277e-73 2
## MGST3.1 1.202772e-77 -2.3332953 0.027 0.424 4.402387e-73 2
## ZNF706 1.253420e-77 -1.2015806 0.077 0.519 4.587768e-73 2
## CORO1B.2 1.368136e-77 -1.4373234 0.123 0.581 5.007651e-73 2
## TMEM59.2 3.074493e-77 -0.9277763 0.128 0.606 1.125326e-72 2
## POLR1D.2 4.033366e-77 -1.2944050 0.081 0.525 1.476292e-72 2
## RBM8A.1 6.254450e-77 -1.0765078 0.087 0.534 2.289254e-72 2
## ATP5MC1 7.181902e-77 -2.2386824 0.029 0.424 2.628720e-72 2
## TMED9.1 7.393430e-77 -1.1566037 0.092 0.544 2.706143e-72 2
## BLOC1S1.1 1.752647e-76 -1.1091484 0.093 0.547 6.415039e-72 2
## EIF4A1 1.884834e-76 -0.9222852 0.302 0.815 6.898869e-72 2
## EIF3M.1 2.082073e-76 -1.0111609 0.135 0.618 7.620803e-72 2
## MRPS24 2.149026e-76 -2.2318902 0.027 0.420 7.865865e-72 2
## ANP32A.1 2.519238e-76 -1.1916620 0.099 0.552 9.220916e-72 2
## ZNHIT1.2 2.912079e-76 -1.5645967 0.057 0.474 1.065879e-71 2
## VDAC2 3.073462e-76 -1.1959502 0.072 0.505 1.124949e-71 2
## MRPL41.1 3.295635e-76 -1.2012738 0.054 0.471 1.206268e-71 2
## LAMTOR2.1 9.396618e-76 -2.2110429 0.030 0.422 3.439350e-71 2
## CCT7 1.257135e-75 -1.6892357 0.053 0.464 4.601366e-71 2
## LIME1.2 1.274474e-75 -1.0001740 0.149 0.628 4.664829e-71 2
## ECH1.2 2.013759e-75 -0.9971309 0.095 0.543 7.370760e-71 2
## BSG.2 2.969913e-75 -0.8890805 0.140 0.622 1.087048e-70 2
## TOMM5 3.392504e-75 -2.0299136 0.042 0.442 1.241724e-70 2
## TRAPPC6A.2 3.448893e-75 -1.2030463 0.080 0.514 1.262364e-70 2
## REEP5.2 4.493964e-75 -1.1310693 0.117 0.581 1.644881e-70 2
## AP2S1.1 5.050998e-75 -1.2569955 0.077 0.507 1.848766e-70 2
## VIM.2 6.124706e-75 -0.9457437 0.475 0.894 2.241765e-70 2
## C11orf58.1 6.474284e-75 -0.9626298 0.092 0.540 2.369718e-70 2
## GTF2A2.1 8.068110e-75 -1.9164413 0.038 0.433 2.953090e-70 2
## GIMAP7.2 9.474357e-75 -0.9680769 0.236 0.752 3.467804e-70 2
## NUTF2 9.800252e-75 -1.6020328 0.053 0.462 3.587088e-70 2
## APH1A.1 1.651084e-74 -2.2441777 0.035 0.426 6.043296e-70 2
## MRPS34 2.158548e-74 -1.0909527 0.083 0.516 7.900717e-70 2
## POLR2E.2 2.477107e-74 -1.2663457 0.066 0.487 9.066708e-70 2
## LSM4.1 3.714942e-74 -1.8101240 0.045 0.445 1.359743e-69 2
## S1PR4.2 3.878994e-74 -1.1390697 0.129 0.589 1.419789e-69 2
## LAMTOR1.1 4.474550e-74 -1.2827762 0.063 0.483 1.637775e-69 2
## RNF181.1 4.721308e-74 -1.7172631 0.039 0.435 1.728093e-69 2
## ATP5MPL 4.740730e-74 -1.5707699 0.051 0.455 1.735202e-69 2
## ATRAID.2 5.244850e-74 -1.3407269 0.048 0.453 1.919720e-69 2
## SRI.2 5.717913e-74 -1.1595821 0.089 0.527 2.092870e-69 2
## CALR.2 8.302266e-74 -0.9006223 0.239 0.750 3.038795e-69 2
## MICOS13.2 8.826577e-74 -1.3819045 0.059 0.471 3.230704e-69 2
## TMEM219.1 1.000083e-73 -2.4090584 0.027 0.409 3.660502e-69 2
## PPP1R18.2 1.093771e-73 -0.8864612 0.137 0.611 4.003420e-69 2
## RNF213.1 1.568494e-73 2.1219013 0.680 0.658 5.741000e-69 2
## CCT2 1.768283e-73 -1.3161566 0.087 0.518 6.472270e-69 2
## ALYREF.1 1.828434e-73 -1.8504788 0.054 0.456 6.692434e-69 2
## PSMD4.1 3.569792e-73 -1.4243846 0.069 0.485 1.306615e-68 2
## NDUFA6.1 4.615698e-73 -1.5697909 0.060 0.467 1.689438e-68 2
## PSMA4.1 6.552727e-73 -2.0761382 0.036 0.422 2.398429e-68 2
## SNRPF 9.554552e-73 -1.7692165 0.047 0.441 3.497157e-68 2
## MRPL54.1 1.268158e-72 -1.0928839 0.060 0.473 4.641710e-68 2
## SIVA1.1 1.669646e-72 -2.0332300 0.042 0.431 6.111238e-68 2
## TPM3.1 1.881264e-72 -0.8267328 0.323 0.834 6.885801e-68 2
## PRKCA.1 2.777877e-72 2.5932027 0.630 0.536 1.016759e-67 2
## TMEM14B 3.911013e-72 -0.9102976 0.090 0.524 1.431509e-67 2
## C12orf75.2 4.302584e-72 -1.4232742 0.075 0.491 1.574832e-67 2
## CACYBP.1 4.748857e-72 -1.6084869 0.063 0.469 1.738177e-67 2
## SELENOF.2 5.680925e-72 -0.8862009 0.117 0.572 2.079332e-67 2
## PDCD6.1 1.017698e-71 -0.9006696 0.072 0.491 3.724979e-67 2
## ELOC.2 2.701522e-71 -1.9087395 0.038 0.421 9.888112e-67 2
## FKBP8.2 2.911645e-71 -0.7939386 0.161 0.646 1.065720e-66 2
## MRPS15.1 2.991635e-71 -1.5547667 0.050 0.443 1.094998e-66 2
## VAMP2.2 3.696048e-71 -1.0046511 0.135 0.597 1.352828e-66 2
## RAB5IF 4.692651e-71 -1.5193385 0.051 0.443 1.717604e-66 2
## POLR2G.2 4.698015e-71 -2.1190418 0.032 0.407 1.719567e-66 2
## JPT1.1 5.222870e-71 -0.7149157 0.102 0.541 1.911675e-66 2
## CHURC1.2 5.289510e-71 -1.2088639 0.090 0.518 1.936066e-66 2
## TKT.1 5.317589e-71 -1.1126165 0.086 0.509 1.946344e-66 2
## NDUFB6 6.596623e-71 -1.5673554 0.050 0.443 2.414496e-66 2
## SUMO1.1 6.662230e-71 -0.8886792 0.081 0.503 2.438510e-66 2
## KDELR1.1 1.088134e-70 -1.8024667 0.059 0.457 3.982788e-66 2
## AURKAIP1.1 1.105456e-70 -1.2151266 0.083 0.505 4.046188e-66 2
## PAIP2.2 1.406979e-70 -0.9584368 0.105 0.542 5.149825e-66 2
## CTDNEP1.2 2.227746e-70 -1.3989183 0.071 0.480 8.153996e-66 2
## MANF.1 2.800864e-70 -1.8846684 0.047 0.433 1.025172e-65 2
## NME1.1 3.071670e-70 -2.0126824 0.035 0.411 1.124293e-65 2
## NDUFB7.1 4.058056e-70 -1.0287226 0.083 0.502 1.485330e-65 2
## MRPL20 4.774926e-70 -1.5229974 0.048 0.436 1.747719e-65 2
## TRAPPC5.1 4.864446e-70 -0.7786818 0.092 0.520 1.780485e-65 2
## EID1.2 5.055075e-70 -0.7410468 0.155 0.633 1.850259e-65 2
## EIF3D.1 5.104680e-70 -0.9963907 0.108 0.544 1.868415e-65 2
## MAGOH.2 5.380154e-70 -1.3879561 0.060 0.460 1.969244e-65 2
## CAPG.1 7.938833e-70 -1.2177877 0.116 0.548 2.905772e-65 2
## PSMB2.1 1.502665e-69 -1.5053248 0.062 0.460 5.500054e-65 2
## ARHGDIA.1 1.665350e-69 -0.9023213 0.152 0.616 6.095513e-65 2
## HOPX.2 1.859101e-69 -1.7465183 0.081 0.479 6.804683e-65 2
## HNRNPK.2 1.925468e-69 -0.7516734 0.218 0.733 7.047597e-65 2
## PPP1R14B.2 2.008042e-69 -1.7119979 0.059 0.450 7.349837e-65 2
## TMEM230.1 2.333330e-69 -1.2560416 0.062 0.461 8.540454e-65 2
## SNRPE.1 2.777080e-69 -1.7155877 0.039 0.415 1.016467e-64 2
## ITM2A.2 2.898547e-69 -1.1585657 0.083 0.492 1.060926e-64 2
## ALDOA 3.164991e-69 -2.1098615 0.038 0.410 1.158450e-64 2
## SET 3.335403e-69 -0.7507504 0.206 0.710 1.220824e-64 2
## BCL2.1 6.848196e-69 2.3245423 0.672 0.647 2.506577e-64 2
## TMED2.1 7.385542e-69 -1.7390096 0.047 0.429 2.703256e-64 2
## PSMB4.2 9.041553e-69 -1.4235436 0.069 0.470 3.309389e-64 2
## SMIM26.2 1.118057e-68 -2.0541093 0.027 0.391 4.092310e-64 2
## OTUB1.2 1.970733e-68 -1.0632388 0.087 0.504 7.213278e-64 2
## SKP1.2 2.143786e-68 -0.7085925 0.224 0.734 7.846685e-64 2
## ANXA5.2 2.379556e-68 -1.4750444 0.068 0.463 8.709652e-64 2
## DNPH1 5.597890e-68 -1.6588281 0.044 0.419 2.048940e-63 2
## PA2G4.1 6.463248e-68 -0.8973937 0.096 0.519 2.365678e-63 2
## STOML2 7.094219e-68 -2.2968560 0.026 0.384 2.596626e-63 2
## LAGE3.1 8.077393e-68 -2.9802590 0.017 0.367 2.956487e-63 2
## PTGES3 9.269781e-68 -0.7520871 0.183 0.673 3.392925e-63 2
## ANXA11.2 1.013416e-67 -0.6812420 0.171 0.655 3.709304e-63 2
## ARF1.2 1.060679e-67 -0.6959996 0.217 0.723 3.882299e-63 2
## MT-ND2.2 1.194483e-67 0.6619370 0.856 0.926 4.372045e-63 2
## HSP90AB1 1.286196e-67 -0.8816082 0.361 0.836 4.707734e-63 2
## ALOX5AP.2 1.349012e-67 -1.5788196 0.089 0.491 4.937656e-63 2
## TMEM14C 1.435820e-67 -2.0675460 0.033 0.397 5.255389e-63 2
## PSMC5.1 1.464869e-67 -0.9782891 0.084 0.498 5.361713e-63 2
## POLD4.2 1.537051e-67 -0.8480424 0.110 0.541 5.625913e-63 2
## DHRS7.2 1.716607e-67 -1.4263211 0.056 0.439 6.283123e-63 2
## PDCD5 2.916974e-67 -1.1869612 0.053 0.435 1.067671e-62 2
## PSMG2 3.800672e-67 -1.5036968 0.051 0.433 1.391122e-62 2
## MRPL34.1 4.165972e-67 -1.5374229 0.047 0.422 1.524829e-62 2
## RANBP1.2 4.421732e-67 -1.4799478 0.060 0.444 1.618443e-62 2
## ETHE1.1 4.472464e-67 -1.3562998 0.051 0.430 1.637011e-62 2
## MT2A.1 4.859452e-67 -1.4350515 0.075 0.468 1.778657e-62 2
## IGBP1.2 6.122584e-67 -1.3200403 0.066 0.458 2.240988e-62 2
## MRPL43 6.165004e-67 -2.2437075 0.032 0.392 2.256515e-62 2
## ETFB.1 7.656454e-67 -1.1571071 0.077 0.481 2.802415e-62 2
## HNRNPA0 8.300492e-67 -0.9353248 0.113 0.543 3.038146e-62 2
## LSM6.1 8.414319e-67 -1.6400936 0.039 0.407 3.079809e-62 2
## ARL6IP5.2 9.180805e-67 -0.6802410 0.289 0.801 3.360358e-62 2
## RNPS1 9.204188e-67 -1.1614897 0.084 0.492 3.368917e-62 2
## SCP2.1 1.110813e-66 -0.8739859 0.074 0.474 4.065798e-62 2
## RNF167.2 1.187489e-66 -0.8818237 0.119 0.551 4.346448e-62 2
## CIAO2A.1 1.212185e-66 -1.7414503 0.039 0.407 4.436839e-62 2
## MYDGF.1 1.480267e-66 -1.1929517 0.092 0.501 5.418073e-62 2
## SRM 2.794932e-66 -1.3696218 0.062 0.446 1.023001e-61 2
## MRPL57 2.941660e-66 -0.9123093 0.065 0.456 1.076706e-61 2
## CCT3.1 3.746372e-66 -0.8682366 0.113 0.541 1.371247e-61 2
## VDAC1.1 4.174832e-66 -1.4947751 0.057 0.435 1.528072e-61 2
## REXO2.2 5.309402e-66 -2.0200643 0.030 0.387 1.943347e-61 2
## H2AFV 5.430313e-66 -1.0077833 0.161 0.615 1.987603e-61 2
## PRMT1 7.752003e-66 -1.3829446 0.050 0.423 2.837388e-61 2
## GADD45GIP1.1 9.980028e-66 -1.0657380 0.057 0.438 3.652890e-61 2
## PKM.1 1.463833e-65 -0.9735233 0.153 0.600 5.357923e-61 2
## RBIS.1 2.303596e-65 -1.9293117 0.038 0.398 8.431624e-61 2
## MIEN1.1 3.212274e-65 -1.3109840 0.044 0.410 1.175757e-60 2
## HMGB1 5.300120e-65 -0.8862075 0.355 0.847 1.939950e-60 2
## NDUFV2.1 5.928554e-65 -0.8060923 0.170 0.630 2.169969e-60 2
## RNF7.2 8.023707e-65 -1.1634446 0.063 0.447 2.936837e-60 2
## PLSCR3.1 8.607058e-65 -1.4219510 0.063 0.444 3.150355e-60 2
## RHOG.2 8.814791e-65 -1.0861205 0.105 0.521 3.226390e-60 2
## ZNRD1.1 8.875168e-65 -1.3848611 0.041 0.403 3.248489e-60 2
## MRPL14 1.700796e-64 -2.6707989 0.018 0.357 6.225252e-60 2
## COPS9.2 2.350565e-64 -1.8690194 0.024 0.369 8.603538e-60 2
## ILF2.1 2.824289e-64 -1.1087951 0.069 0.455 1.033746e-59 2
## AP2M1.2 2.840182e-64 -0.9421347 0.089 0.491 1.039564e-59 2
## AIP.2 3.200027e-64 -1.0382690 0.095 0.502 1.171274e-59 2
## CHMP2A.1 3.237644e-64 -1.2238871 0.054 0.428 1.185043e-59 2
## ADH5.2 3.531853e-64 -1.4649663 0.050 0.418 1.292729e-59 2
## DUT.2 3.545910e-64 -1.2766981 0.084 0.472 1.297874e-59 2
## GIMAP1.2 3.572629e-64 -0.9569162 0.105 0.516 1.307654e-59 2
## SNRPD1.1 3.602696e-64 -1.5070263 0.059 0.431 1.318659e-59 2
## CMTM3.1 3.641725e-64 -1.3121905 0.068 0.449 1.332944e-59 2
## PHB 3.942775e-64 -1.6870104 0.053 0.419 1.443134e-59 2
## EIF6.1 4.055841e-64 -1.4940931 0.033 0.387 1.484519e-59 2
## APEX1 4.534563e-64 -1.5508985 0.053 0.421 1.659741e-59 2
## XRCC6.1 5.098962e-64 -0.6694034 0.138 0.582 1.866322e-59 2
## SPCS2.1 5.223892e-64 -0.8431699 0.119 0.545 1.912049e-59 2
## UBE2B.2 1.689969e-63 -1.1827393 0.068 0.449 6.185626e-59 2
## TUBA4A 1.734927e-63 -0.7001193 0.129 0.559 6.350179e-59 2
## ACP1.2 2.420289e-63 -1.6294399 0.041 0.398 8.858742e-59 2
## RWDD1.1 3.401591e-63 -1.2709483 0.059 0.431 1.245050e-58 2
## SHMT2 3.565980e-63 -2.4377051 0.027 0.369 1.305220e-58 2
## TMEM123.1 5.044239e-63 -0.6980448 0.215 0.698 1.846292e-58 2
## DCXR.2 5.616425e-63 -1.0254557 0.066 0.444 2.055724e-58 2
## ORAI1.2 5.648134e-63 -1.1067636 0.069 0.449 2.067330e-58 2
## CLPP 5.918251e-63 -1.3916567 0.045 0.403 2.166198e-58 2
## YPEL3.2 6.389120e-63 -0.7271459 0.155 0.599 2.338546e-58 2
## UBE2V1.2 1.228998e-62 -0.6856973 0.105 0.514 4.498378e-58 2
## PSMD13.1 1.514948e-62 -1.0877984 0.071 0.450 5.545013e-58 2
## NDUFAF3.1 1.612363e-62 -1.2354920 0.053 0.417 5.901570e-58 2
## IFI27L2.1 2.459178e-62 -2.0831808 0.026 0.363 9.001083e-58 2
## MAP2K2.2 2.513242e-62 -0.6292769 0.129 0.560 9.198967e-58 2
## PSMC4.1 2.869635e-62 -1.4313983 0.059 0.424 1.050344e-57 2
## NAP1L1.1 3.554959e-62 -0.7032711 0.194 0.660 1.301186e-57 2
## CAP1.2 3.608491e-62 -0.5424826 0.221 0.719 1.320780e-57 2
## ISG20.1 3.738304e-62 -0.7803135 0.143 0.576 1.368294e-57 2
## MBNL1.1 4.280370e-62 1.4980047 0.747 0.852 1.566701e-57 2
## PUF60.2 5.014637e-62 -1.5269680 0.053 0.413 1.835458e-57 2
## METTL26 5.343710e-62 -0.7215288 0.074 0.457 1.955905e-57 2
## ANXA2.2 5.625074e-62 -0.7669924 0.147 0.580 2.058890e-57 2
## MRPL11 7.009707e-62 -1.9035776 0.027 0.365 2.565693e-57 2
## CTSD.1 9.499587e-62 -0.7426876 0.141 0.575 3.477039e-57 2
## C1orf43 1.071124e-61 -0.9095951 0.074 0.455 3.920528e-57 2
## SSNA1.1 1.114545e-61 -0.6182579 0.086 0.477 4.079458e-57 2
## PSMD9.1 1.202204e-61 -1.0963719 0.051 0.412 4.400307e-57 2
## LIMD2.2 1.424950e-61 -0.7786390 0.362 0.844 5.215601e-57 2
## NBDY.1 2.202133e-61 -1.1214340 0.077 0.456 8.060247e-57 2
## MTPN.2 2.514134e-61 -1.0205373 0.087 0.477 9.202232e-57 2
## EIF1.2 2.753743e-61 -0.6828390 0.546 0.909 1.007925e-56 2
## RAD23A 2.796870e-61 -0.8044322 0.107 0.513 1.023711e-56 2
## IMP3.1 5.312728e-61 -0.9624505 0.084 0.470 1.944565e-56 2
## PTP4A2.2 5.633764e-61 -0.5919649 0.179 0.640 2.062070e-56 2
## FIBP.1 7.259934e-61 -1.6443279 0.035 0.376 2.657281e-56 2
## NKG7.2 1.051395e-60 -2.0233834 0.208 0.582 3.848317e-56 2
## SNRPB2.2 1.226499e-60 -1.9340969 0.051 0.402 4.489232e-56 2
## VDAC3 1.826614e-60 -0.8869415 0.068 0.437 6.685773e-56 2
## GTF3C6.1 2.831401e-60 -1.3758339 0.045 0.394 1.036350e-55 2
## CD74.1 2.908877e-60 -0.8078317 0.459 0.874 1.064707e-55 2
## MPG.2 2.961824e-60 -1.5190017 0.041 0.384 1.084087e-55 2
## BIN1.2 3.599465e-60 -0.7279435 0.117 0.528 1.317476e-55 2
## MAT2B.2 3.778687e-60 -0.9790314 0.090 0.476 1.383075e-55 2
## NPC2.1 4.965643e-60 -2.0495869 0.030 0.363 1.817525e-55 2
## HADHA.1 6.117861e-60 -0.5884326 0.138 0.564 2.239259e-55 2
## TMBIM4.1 6.505921e-60 -0.6966928 0.119 0.530 2.381297e-55 2
## TXN2 6.943153e-60 -1.7004109 0.044 0.387 2.541333e-55 2
## PSMC3.1 6.976440e-60 -2.1374253 0.033 0.366 2.553517e-55 2
## SDHD 1.012074e-59 -1.2294690 0.063 0.426 3.704394e-55 2
## SUPT4H1.1 1.044977e-59 -2.0025717 0.027 0.355 3.824823e-55 2
## NDUFS3.1 1.428779e-59 -1.7393242 0.029 0.359 5.229617e-55 2
## STMP1.1 1.512195e-59 -2.2673828 0.026 0.352 5.534937e-55 2
## SSB 1.529819e-59 -1.0721314 0.080 0.454 5.599442e-55 2
## IDH2.2 1.890189e-59 -0.8442335 0.117 0.521 6.918470e-55 2
## TMEM256.1 2.351238e-59 -3.1865653 0.011 0.322 8.606002e-55 2
## CNPY2.1 2.452383e-59 -0.8445077 0.083 0.461 8.976212e-55 2
## BUD31.1 2.518603e-59 -1.4579551 0.048 0.395 9.218591e-55 2
## RPS19BP1 2.634758e-59 -0.9894745 0.060 0.419 9.643741e-55 2
## NDUFB5.2 2.813768e-59 -1.1205935 0.060 0.418 1.029895e-54 2
## FAM174C 2.869389e-59 -1.5990813 0.033 0.367 1.050254e-54 2
## LSM3.1 3.483346e-59 -2.5153407 0.015 0.331 1.274974e-54 2
## PIN1.1 3.768954e-59 -0.9384837 0.075 0.444 1.379512e-54 2
## HMGN3.1 5.275331e-59 -1.4669398 0.044 0.386 1.930877e-54 2
## ZNF428.1 5.600182e-59 -1.1097535 0.050 0.398 2.049778e-54 2
## CSK.1 6.271292e-59 -0.5906376 0.173 0.618 2.295418e-54 2
## PRDX3.1 6.433092e-59 -1.3282941 0.054 0.403 2.354640e-54 2
## MPC2 7.485389e-59 -1.6999663 0.039 0.375 2.739802e-54 2
## CTSC.2 1.067940e-58 -0.7324420 0.203 0.654 3.908872e-54 2
## PPP2R1A.1 1.069624e-58 -0.8946039 0.104 0.499 3.915038e-54 2
## TRADD.1 1.105451e-58 -0.8364963 0.089 0.471 4.046172e-54 2
## LSM5.2 1.147969e-58 -2.3792600 0.018 0.334 4.201795e-54 2
## CALM2.1 1.576560e-58 -0.3383008 0.214 0.686 5.770525e-54 2
## CCT8 1.775080e-58 -0.6505221 0.137 0.555 6.497149e-54 2
## COX14 2.140484e-58 -1.6378846 0.035 0.366 7.834600e-54 2
## NUDT5.1 2.237662e-58 -1.3783523 0.057 0.407 8.190292e-54 2
## CCT4 2.886106e-58 -0.9374315 0.123 0.526 1.056372e-53 2
## EMC4.1 3.247200e-58 -1.5623421 0.036 0.368 1.188540e-53 2
## PYURF.1 3.398586e-58 -1.6100343 0.035 0.365 1.243950e-53 2
## RPN2.1 5.712409e-58 -0.6527034 0.159 0.593 2.090856e-53 2
## SYNGR2 7.464240e-58 -1.0728537 0.066 0.425 2.732061e-53 2
## DEK.2 7.769405e-58 -0.7782867 0.132 0.536 2.843758e-53 2
## HLA-DPA1.2 7.797291e-58 -1.3496178 0.075 0.434 2.853965e-53 2
## SELENOK.1 8.285048e-58 -0.8094186 0.092 0.469 3.032493e-53 2
## HSD17B10.1 1.007264e-57 -1.6838756 0.038 0.368 3.686789e-53 2
## SUMO3.1 1.061153e-57 -1.5509341 0.045 0.380 3.884031e-53 2
## SCAND1.1 1.665605e-57 -0.4685331 0.102 0.495 6.096448e-53 2
## TMEM179B.1 1.875723e-57 -1.4397426 0.033 0.359 6.865521e-53 2
## SELENOT.1 1.988466e-57 -0.8091756 0.086 0.459 7.278183e-53 2
## TXNL1.1 2.029203e-57 -1.2884356 0.051 0.393 7.427287e-53 2
## AL138963.4.1 2.072944e-57 -1.0949899 0.044 0.378 7.587390e-53 2
## RNF187.1 2.840941e-57 -1.2021208 0.063 0.414 1.039841e-52 2
## HINT2 3.717165e-57 -1.4997251 0.035 0.361 1.360557e-52 2
## NAXE 4.103890e-57 -2.4340923 0.015 0.322 1.502106e-52 2
## GSTO1 6.899189e-57 -1.5461082 0.039 0.369 2.525241e-52 2
## PSMA3.1 7.432513e-57 -0.9784621 0.083 0.449 2.720448e-52 2
## ADRM1.1 7.444913e-57 -0.9454417 0.068 0.423 2.724987e-52 2
## TRAPPC2L.1 9.888311e-57 -1.7495597 0.027 0.345 3.619320e-52 2
## POLR2K.1 1.057180e-56 -1.3472396 0.039 0.368 3.869489e-52 2
## UBE2N.1 1.247801e-56 -0.7935175 0.108 0.494 4.567200e-52 2
## PPP1R7.1 1.338403e-56 -1.2479117 0.044 0.376 4.898821e-52 2
## RPN1 1.744424e-56 -0.8483944 0.107 0.491 6.384941e-52 2
## ENY2 1.753928e-56 -1.0873067 0.042 0.373 6.419728e-52 2
## COPS6.1 2.119811e-56 -0.9517188 0.048 0.384 7.758932e-52 2
## SDF2L1.1 2.212088e-56 -1.1869453 0.060 0.406 8.096684e-52 2
## ADSL 2.386566e-56 -0.8392265 0.071 0.425 8.735308e-52 2
## RGS19.2 2.565115e-56 -0.9975114 0.066 0.418 9.388833e-52 2
## RAB5C.1 2.604688e-56 -0.9817264 0.071 0.426 9.533681e-52 2
## STUB1.2 2.728802e-56 -0.7999792 0.068 0.421 9.987961e-52 2
## OXA1L.1 2.760033e-56 -0.7431745 0.084 0.451 1.010227e-51 2
## SNF8 2.772721e-56 -1.5072890 0.041 0.368 1.014871e-51 2
## R3HDM4.2 3.000112e-56 -1.0344076 0.066 0.418 1.098101e-51 2
## VKORC1.1 3.772631e-56 -1.9596905 0.030 0.347 1.380859e-51 2
## RPA3.2 3.940378e-56 -2.1017195 0.026 0.338 1.442257e-51 2
## FLT3LG.1 4.220513e-56 -0.4630858 0.224 0.693 1.544792e-51 2
## XBP1.2 4.748745e-56 -0.5574158 0.170 0.607 1.738136e-51 2
## MMADHC.1 5.585375e-56 -1.3664221 0.059 0.400 2.044359e-51 2
## ABHD14B.2 6.684553e-56 -1.6172099 0.044 0.372 2.446680e-51 2
## DNAJC8.1 8.713584e-56 -0.8317473 0.071 0.425 3.189346e-51 2
## NDUFS6.1 9.149164e-56 -1.0343741 0.068 0.418 3.348777e-51 2
## SPCS3.1 9.279853e-56 -0.6925535 0.126 0.524 3.396612e-51 2
## COA3.1 1.028302e-55 -2.0081124 0.023 0.331 3.763790e-51 2
## AUP1.2 1.229028e-55 -0.5351964 0.077 0.435 4.498490e-51 2
## MLF2.1 1.441317e-55 -1.2102404 0.063 0.408 5.275509e-51 2
## TMEM35B.2 1.478038e-55 -1.5420345 0.041 0.364 5.409913e-51 2
## EIF2S3.2 1.714434e-55 -0.7017764 0.120 0.513 6.275172e-51 2
## KXD1 1.818523e-55 -1.3195481 0.047 0.377 6.656158e-51 2
## PTRHD1.1 2.053906e-55 -1.8921352 0.026 0.335 7.517708e-51 2
## ACAA2.1 2.764776e-55 -0.6591662 0.107 0.489 1.011963e-50 2
## NONO 3.384089e-55 -0.6468042 0.117 0.508 1.238644e-50 2
## NENF.1 4.821009e-55 -2.2338282 0.020 0.323 1.764586e-50 2
## MAZ.1 6.348907e-55 -1.0993962 0.075 0.426 2.323827e-50 2
## ARL6IP1.1 6.968786e-55 -0.5603113 0.170 0.598 2.550715e-50 2
## GSTK1.1 7.994798e-55 -0.5195257 0.299 0.795 2.926256e-50 2
## LSP1.2 8.275699e-55 -0.6075378 0.328 0.831 3.029071e-50 2
## SHISA5.2 8.388748e-55 -0.9018679 0.096 0.465 3.070449e-50 2
## MT-ND6.2 1.063904e-54 0.1849647 0.806 0.839 3.894101e-50 2
## SERPINB1.2 1.183564e-54 -0.5970032 0.117 0.504 4.332081e-50 2
## SF3B2.1 1.225652e-54 -0.5605458 0.177 0.611 4.486130e-50 2
## SH3BGRL.2 1.436596e-54 -0.5916804 0.141 0.552 5.258229e-50 2
## CD8B.2 1.509009e-54 -1.3321226 0.146 0.505 5.523276e-50 2
## CHMP4A.1 1.533796e-54 -0.5454546 0.095 0.464 5.613999e-50 2
## PDCD2 1.603641e-54 -1.3569984 0.042 0.365 5.869647e-50 2
## NUCB2.1 2.050876e-54 -1.0913471 0.065 0.406 7.506616e-50 2
## DSTN.2 2.385884e-54 -0.7962825 0.099 0.469 8.732812e-50 2
## DPY30.1 2.510136e-54 -2.4395918 0.017 0.314 9.187599e-50 2
## TCEA1.1 2.763202e-54 -0.5935679 0.129 0.525 1.011387e-49 2
## LSM8.1 3.003734e-54 -0.7841634 0.072 0.421 1.099427e-49 2
## ZYX.2 3.030461e-54 -0.7455466 0.131 0.522 1.109209e-49 2
## MRPL52 3.716539e-54 -1.3279363 0.039 0.357 1.360327e-49 2
## UQCRC1.1 3.775410e-54 -0.7782288 0.060 0.399 1.381875e-49 2
## SUCLG1 4.264258e-54 -1.5169901 0.050 0.375 1.560804e-49 2
## SEC13.1 5.008013e-54 -0.8913713 0.059 0.394 1.833033e-49 2
## PDIA3.1 5.879465e-54 -0.5408395 0.269 0.757 2.152002e-49 2
## MEA1.1 6.365176e-54 -1.9770547 0.027 0.331 2.329782e-49 2
## PSMF1.1 7.308086e-54 -0.8711688 0.081 0.437 2.674906e-49 2
## REX1BD.2 7.321580e-54 -0.2855003 0.137 0.545 2.679845e-49 2
## NOL7 8.345612e-54 -0.8887526 0.089 0.449 3.054661e-49 2
## MRPS12 9.229153e-54 -1.3704539 0.036 0.349 3.378055e-49 2
## PDIA6.1 9.249465e-54 -0.5705773 0.164 0.582 3.385489e-49 2
## AKR1B1.1 9.778840e-54 -1.5497086 0.042 0.361 3.579251e-49 2
## UBE2D2.1 1.040969e-53 -0.3604915 0.205 0.663 3.810154e-49 2
## ARHGAP15 1.076524e-53 1.6034730 0.725 0.853 3.940295e-49 2
## WASHC3.2 1.136817e-53 -0.7143114 0.075 0.423 4.160976e-49 2
## CCT5 1.180544e-53 -0.6506235 0.101 0.471 4.321025e-49 2
## SCAMP2.2 1.374495e-53 -0.8436562 0.092 0.452 5.030926e-49 2
## DCTN2.1 1.388796e-53 -0.9678676 0.072 0.418 5.083272e-49 2
## PYCR2 1.489913e-53 -1.0571748 0.068 0.407 5.453380e-49 2
## CLNS1A 1.712073e-53 -0.9834092 0.069 0.413 6.266530e-49 2
## COX16.1 2.147617e-53 -2.0731791 0.026 0.328 7.860708e-49 2
## TMEM238 2.615632e-53 -0.9627894 0.054 0.384 9.573738e-49 2
## CCNDBP1.2 3.135913e-53 -1.1201728 0.057 0.387 1.147807e-48 2
## SNX17.1 3.404882e-53 -0.8664951 0.086 0.441 1.246255e-48 2
## NSMCE1.1 3.408731e-53 -0.8951331 0.066 0.405 1.247664e-48 2
## CYC1 3.557344e-53 -0.7304086 0.063 0.400 1.302059e-48 2
## FAAP20.1 4.668560e-53 -1.1239117 0.057 0.388 1.708786e-48 2
## LINC02694.2 5.346822e-53 3.0878344 0.466 0.287 1.957044e-48 2
## LAT.1 5.350642e-53 -0.5073531 0.242 0.713 1.958442e-48 2
## CXCR3.2 6.581723e-53 -1.1417159 0.113 0.474 2.409042e-48 2
## PABPN1.1 7.165389e-53 -0.4463639 0.138 0.537 2.622676e-48 2
## PSMD7.1 7.625173e-53 -0.6988956 0.086 0.439 2.790966e-48 2
## SLC9A3R1.2 8.127971e-53 -0.5189600 0.241 0.698 2.975000e-48 2
## PPIH.1 1.010954e-52 -2.3807331 0.015 0.304 3.700295e-48 2
## HLA-F.2 1.097284e-52 -0.4979875 0.174 0.597 4.016277e-48 2
## TIMM13 1.152946e-52 -1.9576247 0.027 0.326 4.220014e-48 2
## FDFT1.1 1.193590e-52 -1.0783597 0.081 0.427 4.368779e-48 2
## URM1.1 1.441085e-52 -1.1585421 0.054 0.379 5.274659e-48 2
## HAX1 1.514008e-52 -0.8349783 0.059 0.388 5.541573e-48 2
## EIF4B.1 1.594556e-52 -0.4176808 0.241 0.717 5.836393e-48 2
## IAH1.1 1.641453e-52 -1.4106741 0.035 0.341 6.008045e-48 2
## KDELR2 1.689453e-52 -1.3892967 0.045 0.361 6.183736e-48 2
## SARNP.1 2.387922e-52 -0.8290135 0.071 0.411 8.740273e-48 2
## TMEM147 2.590443e-52 -1.6171858 0.027 0.326 9.481540e-48 2
## ILK.1 2.776542e-52 -0.5900941 0.092 0.451 1.016270e-47 2
## SELPLG.2 3.557948e-52 -0.3685806 0.223 0.673 1.302280e-47 2
## TEX264.1 3.674565e-52 -1.3255575 0.047 0.361 1.344964e-47 2
## MRPL4 3.790449e-52 -1.0917429 0.042 0.353 1.387380e-47 2
## PTGER2.2 4.533964e-52 -0.7297805 0.126 0.504 1.659522e-47 2
## EMC6.1 4.742626e-52 -1.1647253 0.030 0.331 1.735896e-47 2
## ARPC5L.1 5.375778e-52 -0.6490520 0.113 0.483 1.967642e-47 2
## BRI3 6.483027e-52 -0.6771831 0.090 0.444 2.372918e-47 2
## CLTB.1 8.293523e-52 -0.8721347 0.069 0.406 3.035595e-47 2
## CNPY3.1 9.396353e-52 -0.8349598 0.074 0.414 3.439253e-47 2
## UBALD2.2 1.009607e-51 -0.3594773 0.202 0.637 3.695363e-47 2
## CISD3.2 1.105328e-51 -1.8142646 0.027 0.322 4.045722e-47 2
## ZMAT2.1 1.158561e-51 -1.2001397 0.054 0.376 4.240567e-47 2
## CBX3.1 1.291615e-51 -0.5582452 0.120 0.499 4.727569e-47 2
## TMEM160.1 1.392189e-51 -0.7092351 0.077 0.417 5.095691e-47 2
## GDI2 1.444431e-51 -0.5257176 0.140 0.530 5.286905e-47 2
## RAB1B.2 1.599388e-51 -0.5500119 0.104 0.470 5.854080e-47 2
## RAB11B.2 1.905125e-51 -0.9756723 0.077 0.415 6.973137e-47 2
## SERBP1.1 2.103207e-51 -0.5520834 0.198 0.636 7.698157e-47 2
## COPZ1.1 2.312456e-51 -0.9632706 0.069 0.402 8.464053e-47 2
## UBL7.1 2.333431e-51 -1.8306876 0.029 0.324 8.540824e-47 2
## CKLF.2 2.487369e-51 -0.5660032 0.135 0.521 9.104268e-47 2
## VPS29 3.595944e-51 -0.4563596 0.092 0.447 1.316187e-46 2
## TUBA1B.2 4.206328e-51 -1.3967224 0.191 0.580 1.539600e-46 2
## MRPS7 4.363170e-51 -1.4119704 0.036 0.338 1.597008e-46 2
## PDE3B.2 5.010003e-51 1.5893872 0.708 0.768 1.833761e-46 2
## RER1.1 5.291790e-51 -0.6144921 0.099 0.455 1.936901e-46 2
## EMD.1 6.640510e-51 -1.0956233 0.045 0.355 2.430560e-46 2
## HSPB1.1 7.790128e-51 -1.4405539 0.038 0.339 2.851343e-46 2
## GET3.2 1.041054e-50 -1.0908188 0.053 0.369 3.810465e-46 2
## HPRT1.1 1.148544e-50 -1.6021780 0.038 0.338 4.203900e-46 2
## TMBIM6.1 1.250616e-50 -0.4800503 0.244 0.702 4.577506e-46 2
## PCBP2 1.391859e-50 -0.4388996 0.138 0.528 5.094482e-46 2
## SSU72.2 1.548864e-50 -0.5400795 0.108 0.471 5.669154e-46 2
## BZW1.2 2.053418e-50 -0.4579291 0.135 0.522 7.515920e-46 2
## CDKN2D.2 2.121115e-50 -1.7206931 0.044 0.347 7.763706e-46 2
## CARHSP1.1 2.364769e-50 -1.1924396 0.051 0.362 8.655526e-46 2
## CHCHD5.2 2.365811e-50 -1.5976546 0.024 0.312 8.659340e-46 2
## HNRNPA3 2.459233e-50 -0.4985997 0.256 0.711 9.001284e-46 2
## ICAM2.2 2.590035e-50 -0.7020114 0.083 0.422 9.480046e-46 2
## MRPS36.1 4.126187e-50 -1.2400979 0.035 0.330 1.510267e-45 2
## THOC7 4.166389e-50 -1.8020019 0.033 0.327 1.524982e-45 2
## ABHD17A.2 4.510531e-50 -0.3107214 0.180 0.603 1.650945e-45 2
## FCMR.2 4.837654e-50 -0.8790346 0.080 0.412 1.770678e-45 2
## MFNG.2 5.015399e-50 -0.6394703 0.107 0.467 1.835737e-45 2
## RHOC.2 5.690730e-50 -1.5150155 0.047 0.352 2.082921e-45 2
## PSMB7 5.726080e-50 -1.0349590 0.059 0.375 2.095860e-45 2
## CCDC167.2 5.767518e-50 -2.0332627 0.021 0.304 2.111027e-45 2
## PFDN2 1.006311e-49 -1.0958355 0.056 0.371 3.683299e-45 2
## C14orf119.1 1.029381e-49 -1.7561138 0.032 0.324 3.767740e-45 2
## TNFSF12.1 1.115764e-49 -1.0981028 0.072 0.399 4.083919e-45 2
## AKR7A2.1 1.125811e-49 -2.2345447 0.020 0.299 4.120694e-45 2
## MRPL18 1.247987e-49 -1.3258814 0.038 0.335 4.567883e-45 2
## NAPA.1 1.381470e-49 -0.3276848 0.105 0.464 5.056457e-45 2
## ANP32E.1 1.569720e-49 -0.9879479 0.081 0.414 5.745490e-45 2
## SRP19.2 1.573598e-49 -0.6526964 0.071 0.399 5.759684e-45 2
## HLA-DPB1.2 1.772992e-49 -1.1452621 0.074 0.399 6.489504e-45 2
## CCNG1.1 2.198524e-49 -1.4879114 0.045 0.346 8.047037e-45 2
## PCMT1.1 2.640846e-49 -0.7267067 0.065 0.386 9.666024e-45 2
## IMPDH2.2 2.810641e-49 -1.0669292 0.077 0.405 1.028751e-44 2
## HMGB2.1 2.824997e-49 -1.0701408 0.137 0.500 1.034006e-44 2
## CHCHD1.1 2.869935e-49 -1.9003060 0.023 0.304 1.050454e-44 2
## ROMO1 3.394107e-49 -1.0130411 0.045 0.346 1.242311e-44 2
## FDPS.1 3.454549e-49 -1.1255815 0.057 0.370 1.264434e-44 2
## P4HB.1 3.904755e-49 -0.4913599 0.244 0.690 1.429218e-44 2
## SLC25A39 4.543995e-49 -0.8008842 0.057 0.370 1.663193e-44 2
## RBM42.1 4.987590e-49 -1.0523395 0.062 0.377 1.825558e-44 2
## PDAP1 5.411673e-49 -0.7154708 0.068 0.390 1.980780e-44 2
## SNRPA 7.076386e-49 -0.8508343 0.068 0.387 2.590099e-44 2
## HSP90B1.1 7.821198e-49 -0.5164179 0.286 0.740 2.862715e-44 2
## MRPL21 9.667492e-49 -1.3579480 0.033 0.321 3.538495e-44 2
## VSIR.1 1.114525e-48 -0.6739373 0.080 0.411 4.079384e-44 2
## TRAM1.2 1.238546e-48 -0.4800503 0.161 0.559 4.533326e-44 2
## TXNDC17.2 1.411993e-48 -1.2281797 0.042 0.339 5.168175e-44 2
## GLRX.1 1.429904e-48 -1.8517537 0.024 0.304 5.233735e-44 2
## DOK2.2 1.611226e-48 -0.4978341 0.116 0.479 5.897408e-44 2
## EMC7.1 1.737139e-48 -1.2759608 0.042 0.338 6.358276e-44 2
## BUB3.1 2.037993e-48 -0.2935940 0.155 0.551 7.459460e-44 2
## TAP1.1 2.073514e-48 -0.5075398 0.183 0.593 7.589477e-44 2
## NUDT21.1 2.335688e-48 -0.9142694 0.059 0.369 8.549086e-44 2
## DGCR6L 2.645576e-48 -1.8671936 0.030 0.313 9.683339e-44 2
## CNIH1 2.964441e-48 -1.2874973 0.050 0.352 1.085045e-43 2
## HMGA1.2 4.778651e-48 -1.3191668 0.065 0.377 1.749082e-43 2
## LEPROTL1.1 4.881143e-48 -0.3329146 0.205 0.634 1.786596e-43 2
## SMARCE1 5.516699e-48 -0.8007799 0.056 0.364 2.019222e-43 2
## STRAP.2 5.862777e-48 -0.8144518 0.080 0.407 2.145894e-43 2
## H1FX.1 8.660672e-48 -0.5321929 0.217 0.626 3.169979e-43 2
## MRPL16 1.291832e-47 -1.2292927 0.051 0.352 4.728365e-43 2
## DGUOK.2 1.349076e-47 -0.5683734 0.104 0.450 4.937889e-43 2
## PRKCH.1 1.429596e-47 1.9200083 0.644 0.691 5.232606e-43 2
## AQP3.2 1.586556e-47 -1.2864781 0.068 0.374 5.807114e-43 2
## TMED10.1 1.615450e-47 -0.3294901 0.122 0.486 5.912871e-43 2
## TRAC.2 1.787626e-47 -0.4587018 0.152 0.531 6.543070e-43 2
## ISG15.1 2.045480e-47 -0.5380061 0.102 0.448 7.486867e-43 2
## MRPL17.1 2.257904e-47 -1.8649276 0.023 0.295 8.264382e-43 2
## SRSF5.2 2.483855e-47 -0.4312658 0.304 0.786 9.091406e-43 2
## NUCKS1.2 2.556973e-47 -0.6209946 0.134 0.499 9.359033e-43 2
## PPM1G 2.604087e-47 -0.3398075 0.129 0.497 9.531478e-43 2
## THYN1.1 2.612255e-47 -0.8719653 0.044 0.337 9.561377e-43 2
## RHOF.1 2.680344e-47 -0.3525313 0.122 0.485 9.810593e-43 2
## HSBP1.1 2.719285e-47 -1.6272967 0.026 0.301 9.953127e-43 2
## COMMD3.1 2.768867e-47 -1.3258806 0.039 0.328 1.013461e-42 2
## COMMD7.1 4.230956e-47 -0.5745731 0.092 0.425 1.548615e-42 2
## POLR2F.1 4.534008e-47 -1.4230419 0.032 0.311 1.659537e-42 2
## NEAT1.1 4.597262e-47 2.1370707 0.626 0.642 1.682690e-42 2
## HSPB11.2 4.616358e-47 -0.7722463 0.062 0.371 1.689679e-42 2
## MOSPD3.1 4.664083e-47 -1.4738138 0.032 0.310 1.707148e-42 2
## LAMP1.1 4.861484e-47 -0.7550989 0.096 0.431 1.779400e-42 2
## TRBC1.2 4.987284e-47 -1.1169154 0.087 0.408 1.825446e-42 2
## RPA2 5.064541e-47 -1.1113531 0.048 0.344 1.853723e-42 2
## RPL22L1 5.223966e-47 -0.6047145 0.066 0.379 1.912076e-42 2
## NUDCD2 6.613068e-47 -1.6811257 0.026 0.299 2.420515e-42 2
## LGALS3.2 6.691392e-47 -1.3428062 0.068 0.372 2.449183e-42 2
## DMAC1 6.926103e-47 -1.7165628 0.024 0.295 2.535092e-42 2
## METTL9.1 6.983613e-47 -0.6059648 0.077 0.398 2.556142e-42 2
## TMCO1.1 7.295179e-47 -1.2421752 0.060 0.363 2.670181e-42 2
## CD47.2 8.750317e-47 -0.4159199 0.159 0.549 3.202791e-42 2
## ECHS1.1 9.742846e-47 -1.7120539 0.026 0.299 3.566076e-42 2
## CMPK1 9.976823e-47 -0.5785019 0.092 0.426 3.651717e-42 2
## SARAF.1 1.118485e-46 -0.5235528 0.383 0.846 4.093878e-42 2
## CDC26.1 1.152662e-46 -1.7280685 0.024 0.295 4.218972e-42 2
## GLRX5 1.243719e-46 -1.1264752 0.039 0.324 4.552261e-42 2
## ESD.1 1.526545e-46 -1.2008352 0.032 0.310 5.587460e-42 2
## MESD 1.580961e-46 -1.0448972 0.071 0.382 5.786635e-42 2
## NUCB1.1 1.805466e-46 -0.3838160 0.156 0.542 6.608368e-42 2
## WAS.2 2.094463e-46 -0.1341651 0.195 0.613 7.666153e-42 2
## NDUFA8 2.293572e-46 -1.6774895 0.023 0.291 8.394934e-42 2
## CCDC107.1 2.440818e-46 -0.9076882 0.059 0.360 8.933881e-42 2
## UQCRC2 2.565715e-46 -0.4330197 0.096 0.433 9.391031e-42 2
## ANKRD44 2.708659e-46 1.8115787 0.639 0.720 9.914235e-42 2
## LYPLA2.1 3.586923e-46 -1.0065153 0.054 0.352 1.312886e-41 2
## NDUFS2.2 3.616727e-46 -0.5746309 0.078 0.396 1.323795e-41 2
## TMEM208 3.821430e-46 -1.6225787 0.024 0.293 1.398720e-41 2
## CDC42.2 3.880178e-46 -0.4408882 0.289 0.755 1.420223e-41 2
## SDHAF2.1 4.422393e-46 -1.0770620 0.051 0.344 1.618684e-41 2
## NECAP2.2 6.440679e-46 -0.6673367 0.080 0.398 2.357417e-41 2
## MRPS6.1 8.067709e-46 -0.7007258 0.062 0.365 2.952943e-41 2
## CANX.1 1.247497e-45 -0.3246239 0.221 0.647 4.566087e-41 2
## UFD1 1.435865e-45 -1.0223844 0.056 0.352 5.255553e-41 2
## CYB5R3.1 1.519172e-45 -1.1937118 0.041 0.322 5.560472e-41 2
## IER3IP1.2 1.644039e-45 -1.1840574 0.042 0.326 6.017512e-41 2
## PPP1CC 1.711163e-45 -0.5929603 0.101 0.436 6.263199e-41 2
## NCR3.2 2.046622e-45 -1.8214655 0.045 0.326 7.491047e-41 2
## LSM10 3.705826e-45 -1.3103247 0.032 0.303 1.356406e-40 2
## RNF5.1 4.360398e-45 -1.5395709 0.033 0.306 1.595993e-40 2
## AKR1A1.1 4.481823e-45 -0.6856229 0.054 0.347 1.640437e-40 2
## SYF2.1 4.923787e-45 -0.5252394 0.084 0.403 1.802204e-40 2
## SF3A2.1 4.960711e-45 -0.6296976 0.084 0.403 1.815720e-40 2
## SF3A1.1 6.059667e-45 -0.8191442 0.080 0.394 2.217959e-40 2
## TMUB1 6.581048e-45 -0.8151819 0.053 0.344 2.408795e-40 2
## CCDC85B.2 6.616058e-45 -0.1489495 0.220 0.653 2.421610e-40 2
## EIF1AY 6.800824e-45 -1.3067781 0.038 0.314 2.489237e-40 2
## MRPS18C.1 6.981933e-45 -1.2772339 0.026 0.291 2.555527e-40 2
## SS18L2.1 7.640797e-45 -0.3300541 0.084 0.404 2.796684e-40 2
## SEPTIN1.1 8.033404e-45 -0.2894437 0.259 0.712 2.940387e-40 2
## MRPL13.1 8.053107e-45 -1.3555281 0.026 0.289 2.947598e-40 2
## RNASET2 8.195457e-45 -0.4644413 0.126 0.475 2.999701e-40 2
## COA4 8.507925e-45 -1.8332274 0.021 0.281 3.114071e-40 2
## CNIH4 8.522544e-45 -1.9028789 0.029 0.295 3.119422e-40 2
## PSMD6 9.797205e-45 -1.1198011 0.057 0.350 3.585973e-40 2
## CUEDC2 1.050639e-44 -1.1261460 0.047 0.329 3.845550e-40 2
## BCL7C.2 1.077032e-44 -0.5240737 0.068 0.371 3.942153e-40 2
## TSR3.1 1.080997e-44 -1.4902181 0.039 0.315 3.956665e-40 2
## H2AFY.1 1.182231e-44 -0.4338738 0.134 0.491 4.327203e-40 2
## SLC25A11.2 1.249934e-44 -1.7470344 0.032 0.300 4.575008e-40 2
## TXNIP.2 1.364923e-44 -0.4829759 0.359 0.786 4.995892e-40 2
## RALY 1.468509e-44 -0.2827774 0.191 0.597 5.375035e-40 2
## C12orf10 1.550256e-44 -2.9839437 0.014 0.264 5.674247e-40 2
## JOSD2.1 1.871537e-44 -0.7752561 0.042 0.321 6.850199e-40 2
## MRPS33.1 2.053632e-44 -1.3115515 0.036 0.308 7.516702e-40 2
## IK.1 2.076653e-44 -0.5147507 0.104 0.435 7.600965e-40 2
## GCHFR.2 2.336611e-44 -1.7963135 0.027 0.290 8.552464e-40 2
## HNRNPR 2.980324e-44 -0.5665374 0.149 0.514 1.090858e-39 2
## EGLN2.2 3.219551e-44 -1.1169032 0.048 0.330 1.178420e-39 2
## HNRNPD.1 3.284337e-44 -0.4500222 0.220 0.640 1.202133e-39 2
## PNKD.1 3.349360e-44 -0.5579548 0.053 0.340 1.225933e-39 2
## C9orf78.1 3.513082e-44 -0.2931762 0.147 0.516 1.285858e-39 2
## MRFAP1L1.1 3.757279e-44 -0.7902404 0.051 0.335 1.375239e-39 2
## NAA38.1 3.793188e-44 -0.9618776 0.041 0.315 1.388383e-39 2
## ITPA.1 4.196033e-44 -1.0113254 0.047 0.328 1.535832e-39 2
## MRPS16.1 4.807588e-44 -1.4966911 0.033 0.301 1.759673e-39 2
## ELP5 4.998253e-44 -1.4034652 0.023 0.280 1.829461e-39 2
## RBMX 5.118163e-44 -0.2180058 0.122 0.469 1.873350e-39 2
## CASP4.2 5.433064e-44 -1.1546356 0.050 0.331 1.988610e-39 2
## HSPD1.2 7.385964e-44 -0.5240098 0.195 0.583 2.703410e-39 2
## CTSW.2 7.595849e-44 -1.2438038 0.099 0.406 2.780233e-39 2
## UBXN4.1 8.420211e-44 -0.2274372 0.203 0.618 3.081966e-39 2
## TMEM109.1 8.920654e-44 -1.4059707 0.045 0.322 3.265138e-39 2
## HIKESHI 8.976363e-44 -1.2062249 0.045 0.323 3.285529e-39 2
## PSENEN.1 9.090659e-44 -1.4507657 0.026 0.285 3.327363e-39 2
## SNHG16.1 9.451333e-44 -1.1508901 0.051 0.334 3.459377e-39 2
## UBE2M.1 9.706617e-44 -0.9948984 0.044 0.321 3.552816e-39 2
## NDUFA3.1 9.866285e-44 -0.9261340 0.042 0.317 3.611258e-39 2
## CNPPD1.2 9.931999e-44 -0.7071035 0.066 0.363 3.635310e-39 2
## GOLGA7.1 1.092391e-43 -1.1538742 0.044 0.320 3.998368e-39 2
## NDUFV1.1 1.097820e-43 -0.1325783 0.128 0.482 4.018242e-39 2
## OCIAD1.1 1.122133e-43 -0.4115626 0.122 0.469 4.107231e-39 2
## SLC2A4RG 1.226032e-43 -0.9206469 0.057 0.344 4.487522e-39 2
## MRPL33.1 1.279455e-43 -1.0129052 0.030 0.294 4.683060e-39 2
## SH3BP1.2 1.747365e-43 -0.5687718 0.086 0.398 6.395705e-39 2
## TIMM8B.1 1.830513e-43 -1.6428751 0.018 0.269 6.700045e-39 2
## IFI6.1 2.071776e-43 -1.0935917 0.039 0.309 7.583115e-39 2
## OSTF1.2 2.076891e-43 -0.3142540 0.165 0.540 7.601835e-39 2
## M6PR.1 2.088323e-43 -0.4705488 0.119 0.457 7.643680e-39 2
## CLTA 2.125351e-43 -0.1343783 0.173 0.564 7.779211e-39 2
## MCTS1 2.150649e-43 -1.4393686 0.026 0.283 7.871807e-39 2
## FUNDC2 2.766793e-43 -1.3178772 0.032 0.295 1.012701e-38 2
## THAP11 3.052133e-43 -2.5701865 0.012 0.256 1.117142e-38 2
## MEAF6.1 3.454666e-43 -0.8403968 0.065 0.357 1.264477e-38 2
## PRPF19 3.721924e-43 -1.2344293 0.038 0.305 1.362299e-38 2
## ZDHHC12 4.024228e-43 -2.4841866 0.018 0.266 1.472948e-38 2
## DBNL.1 4.493054e-43 -0.4364326 0.104 0.431 1.644548e-38 2
## TAF9 5.075889e-43 -1.8728715 0.021 0.273 1.857877e-38 2
## CENPX.1 6.771510e-43 -0.7296191 0.053 0.335 2.478508e-38 2
## EVI2B.2 7.065537e-43 -0.5533129 0.087 0.393 2.586128e-38 2
## NDUFB1 7.086073e-43 -1.6116649 0.024 0.277 2.593645e-38 2
## ARL2BP.1 7.248102e-43 -1.0704887 0.044 0.317 2.652950e-38 2
## TIMM17B 7.627627e-43 -1.6453395 0.021 0.273 2.791864e-38 2
## HLA-E.2 9.115855e-43 -0.5473174 0.438 0.876 3.336585e-38 2
## IDH3G.1 9.393083e-43 -1.0137627 0.045 0.319 3.438056e-38 2
## PSIP1 9.536986e-43 -0.2847300 0.153 0.518 3.490728e-38 2
## ZC3H15.2 1.046528e-42 -0.4722867 0.096 0.415 3.830504e-38 2
## CHMP5.1 1.109464e-42 -1.3950486 0.038 0.304 4.060859e-38 2
## NUDC.1 1.541590e-42 -0.5011526 0.090 0.402 5.642526e-38 2
## PSMB8-AS1.2 1.549113e-42 -0.6241964 0.059 0.344 5.670065e-38 2
## LYAR.2 1.565464e-42 -1.2769073 0.048 0.321 5.729913e-38 2
## CTSA.1 1.631135e-42 -1.4578872 0.041 0.306 5.970281e-38 2
## STX10 1.925267e-42 -1.4551908 0.036 0.300 7.046861e-38 2
## MRPL36 2.158591e-42 -1.6702730 0.020 0.267 7.900873e-38 2
## FKBP11.2 2.227669e-42 -0.8321584 0.093 0.401 8.153715e-38 2
## VASP.1 2.319781e-42 -0.3503465 0.129 0.474 8.490863e-38 2
## TMEM9B.2 2.342824e-42 -1.3884318 0.038 0.302 8.575205e-38 2
## POLE4 3.014477e-42 -2.7907606 0.009 0.245 1.103359e-37 2
## DNAJC19.1 3.387355e-42 -1.5742447 0.026 0.278 1.239840e-37 2
## SUSD3.1 3.460809e-42 -1.1172247 0.041 0.307 1.266725e-37 2
## SMIM19 3.687406e-42 -1.6642712 0.018 0.263 1.349664e-37 2
## METTL5 4.243924e-42 -1.1626136 0.033 0.292 1.553361e-37 2
## MAF1.1 4.761546e-42 -1.0311628 0.051 0.325 1.742821e-37 2
## COMT.1 7.324493e-42 -0.8228040 0.050 0.323 2.680911e-37 2
## PPP1R2 7.925973e-42 -0.3656352 0.110 0.435 2.901065e-37 2
## DNAJA2.1 8.084647e-42 -0.7591722 0.062 0.345 2.959143e-37 2
## LITAF.2 8.843354e-42 -0.4132508 0.128 0.467 3.236844e-37 2
## ARF6.2 9.650735e-42 -0.2510048 0.212 0.624 3.532362e-37 2
## HDAC1 1.051764e-41 -0.3056834 0.138 0.483 3.849665e-37 2
## NDUFA5.1 1.140216e-41 -1.8859272 0.020 0.264 4.173419e-37 2
## TRAPPC3.1 1.199451e-41 -0.8522205 0.051 0.326 4.390231e-37 2
## SNHG32.1 1.278228e-41 -1.5995826 0.020 0.262 4.678571e-37 2
## NIFK 1.509277e-41 -1.1543238 0.033 0.290 5.524254e-37 2
## CARD16.1 1.525650e-41 -1.3971332 0.033 0.289 5.584183e-37 2
## DENND2D.1 1.590259e-41 -0.2041350 0.162 0.534 5.820664e-37 2
## NDUFC1 1.635707e-41 -0.9453708 0.053 0.327 5.987016e-37 2
## FAM162A 1.647439e-41 -1.3014332 0.038 0.298 6.029956e-37 2
## HLA-DRB1.2 1.852464e-41 -0.9054194 0.147 0.477 6.780387e-37 2
## AGTRAP.2 1.854886e-41 -1.1677234 0.050 0.319 6.789255e-37 2
## NAA20 1.938264e-41 -1.2999217 0.032 0.287 7.094435e-37 2
## DCTPP1.2 2.118170e-41 -2.0072904 0.023 0.268 7.752926e-37 2
## CCDC124 2.320025e-41 -1.0169256 0.050 0.320 8.491756e-37 2
## ARL2.2 2.327639e-41 -1.1520450 0.023 0.268 8.519624e-37 2
## SNX6 2.513623e-41 -0.1329947 0.132 0.477 9.200361e-37 2
## COA6 2.745518e-41 -1.4525180 0.030 0.282 1.004914e-36 2
## POLR2H.1 3.165126e-41 -1.1121171 0.036 0.294 1.158499e-36 2
## NUBP2 3.418899e-41 -1.0086804 0.038 0.296 1.251385e-36 2
## PQBP1.1 4.013650e-41 -0.6509746 0.060 0.340 1.469076e-36 2
## CD320 5.559460e-41 -1.7392646 0.030 0.280 2.034874e-36 2
## ATG3 6.436837e-41 -0.6166345 0.060 0.339 2.356011e-36 2
## BLOC1S2.1 6.777174e-41 -1.1096276 0.041 0.301 2.480581e-36 2
## SDHB 9.855958e-41 -0.7729934 0.042 0.303 3.607478e-36 2
## GLRX3 1.032827e-40 -1.1144170 0.039 0.296 3.780352e-36 2
## ZNHIT3.1 1.041474e-40 -1.0071402 0.038 0.295 3.812004e-36 2
## TXNDC12.1 1.063488e-40 -1.0306800 0.051 0.320 3.892577e-36 2
## PHF5A 1.084575e-40 -1.4007045 0.032 0.283 3.969763e-36 2
## RNH1.1 1.156505e-40 -0.5042684 0.099 0.407 4.233038e-36 2
## SDF4.2 1.199031e-40 -0.1545269 0.105 0.424 4.388692e-36 2
## MACF1.2 1.215371e-40 1.8913260 0.592 0.602 4.448500e-36 2
## PRELID3B 1.374069e-40 -1.3312012 0.039 0.297 5.029366e-36 2
## POLR2I 1.374495e-40 -1.3144458 0.017 0.253 5.030926e-36 2
## GBP2.1 1.397501e-40 -0.3846724 0.114 0.433 5.115131e-36 2
## AP1S1.1 1.814107e-40 -1.9994237 0.026 0.270 6.639996e-36 2
## AP1M1 1.901530e-40 -0.4910334 0.066 0.348 6.959982e-36 2
## AHSA1.1 1.903193e-40 -1.2818110 0.041 0.298 6.966069e-36 2
## COPS5.1 2.016031e-40 -0.7433965 0.060 0.336 7.379075e-36 2
## SRP72 2.023474e-40 -0.3807565 0.092 0.396 7.406319e-36 2
## FERMT3.1 2.305668e-40 -0.1960500 0.162 0.525 8.439205e-36 2
## PIM1.2 2.793419e-40 -0.2579003 0.215 0.614 1.022447e-35 2
## H2AFJ.1 2.868011e-40 -0.8178159 0.045 0.307 1.049749e-35 2
## SCAMP3 3.008012e-40 -0.6363780 0.069 0.353 1.100992e-35 2
## FLOT1.2 3.112316e-40 -1.3277451 0.041 0.298 1.139170e-35 2
## CDK4.2 3.249040e-40 -1.1973583 0.039 0.294 1.189214e-35 2
## POP5 3.341208e-40 -0.9737640 0.032 0.281 1.222949e-35 2
## NDUFA9 3.345560e-40 -0.7488497 0.054 0.325 1.224542e-35 2
## FLOT2.2 3.730257e-40 -0.5204173 0.092 0.393 1.365349e-35 2
## PPT1 4.191633e-40 -1.1485795 0.045 0.305 1.534222e-35 2
## CISD1 4.254694e-40 -1.7532582 0.018 0.253 1.557303e-35 2
## TIMM10.1 4.444450e-40 -2.1716173 0.018 0.252 1.626758e-35 2
## ATP6V0D1.1 4.626798e-40 -0.6916692 0.057 0.330 1.693501e-35 2
## GRHPR.1 4.875772e-40 -0.8921989 0.048 0.311 1.784630e-35 2
## MRPL28.1 6.095954e-40 -0.9007554 0.044 0.302 2.231241e-35 2
## RPAIN 6.159818e-40 -0.7863835 0.048 0.311 2.254616e-35 2
## PTDSS1.1 7.598044e-40 -0.7157731 0.081 0.371 2.781036e-35 2
## FBXO7.1 7.654381e-40 -0.2900580 0.089 0.388 2.801657e-35 2
## RCN2.1 7.890338e-40 -1.4397596 0.036 0.287 2.888021e-35 2
## BOLA3.1 8.493731e-40 -1.8983690 0.023 0.261 3.108875e-35 2
## JUN.1 9.415969e-40 1.2036658 0.839 0.870 3.446433e-35 2
## SPAG7.1 1.090546e-39 -1.0219021 0.048 0.309 3.991617e-35 2
## TADA3.1 1.137997e-39 -0.5394908 0.077 0.363 4.165296e-35 2
## ORMDL2.1 1.187876e-39 -1.6490703 0.030 0.274 4.347862e-35 2
## COX17 1.229543e-39 -1.1230417 0.044 0.300 4.500373e-35 2
## SRA1 1.342566e-39 -0.6387409 0.054 0.321 4.914059e-35 2
## EIF1B.1 1.425820e-39 -1.2177965 0.039 0.292 5.218785e-35 2
## PFDN6 1.519300e-39 -1.0133284 0.036 0.286 5.560943e-35 2
## ELOF1.1 1.576117e-39 -2.2497885 0.014 0.242 5.768902e-35 2
## SMIM12.1 1.712301e-39 -1.1675534 0.032 0.277 6.267363e-35 2
## PSMC2.1 1.901003e-39 -1.1259069 0.048 0.308 6.958052e-35 2
## ETFA.1 2.110642e-39 -0.3075914 0.090 0.391 7.725373e-35 2
## IGFLR1.1 2.166155e-39 -1.0180558 0.057 0.324 7.928559e-35 2
## MRPL9 2.401548e-39 -1.4580969 0.030 0.273 8.790146e-35 2
## YWHAH.1 2.675484e-39 -0.6114705 0.093 0.389 9.792808e-35 2
## PAFAH1B3.2 2.901425e-39 -2.5037113 0.011 0.234 1.061980e-34 2
## POLR3GL.1 3.106117e-39 -0.6067523 0.068 0.345 1.136901e-34 2
## HMGN4.2 3.116165e-39 -0.5802583 0.074 0.357 1.140579e-34 2
## MOB1A.1 3.193290e-39 -0.2033302 0.129 0.460 1.168808e-34 2
## ANXA7.1 3.226548e-39 -0.4539485 0.099 0.401 1.180981e-34 2
## KLF2.1 3.865123e-39 -0.6417378 0.195 0.536 1.414712e-34 2
## TMEM126A 4.013322e-39 -1.7188716 0.014 0.240 1.468956e-34 2
## MRPS35 4.560544e-39 -0.9513469 0.044 0.298 1.669250e-34 2
## PFDN4 4.578482e-39 -2.5094751 0.014 0.239 1.675816e-34 2
## VEGFB.2 4.910275e-39 -0.5272472 0.068 0.342 1.797259e-34 2
## SELL.1 5.471311e-39 -1.7099009 0.039 0.285 2.002609e-34 2
## SASH3.1 5.622534e-39 -0.1246686 0.131 0.461 2.057960e-34 2
## SNRPA1.1 5.855426e-39 -0.5242085 0.075 0.357 2.143203e-34 2
## NUDT1.2 7.171510e-39 -0.7380070 0.050 0.309 2.624916e-34 2
## ECI1.1 7.507282e-39 -1.8637913 0.020 0.249 2.747815e-34 2
## FKBP3 8.295044e-39 -0.8669642 0.038 0.286 3.036152e-34 2
## YIF1A.1 8.296623e-39 -1.5400902 0.027 0.265 3.036730e-34 2
## LINC01871.2 1.017887e-38 -0.8378229 0.080 0.361 3.725671e-34 2
## GPS2.2 1.057737e-38 -0.1397820 0.098 0.399 3.871529e-34 2
## CWC15 1.077169e-38 -1.2415125 0.036 0.282 3.942654e-34 2
## ATP6V0E2 1.131153e-38 -0.7831348 0.047 0.302 4.140245e-34 2
## CMC2 1.156292e-38 -0.7919900 0.056 0.319 4.232259e-34 2
## SRSF3 1.317549e-38 -0.1908487 0.289 0.727 4.822494e-34 2
## UBE2A.1 1.623996e-38 -0.6729748 0.065 0.336 5.944152e-34 2
## METTL23.2 1.625717e-38 -1.1659064 0.039 0.286 5.950449e-34 2
## LSM1.1 1.876701e-38 -0.8114710 0.041 0.290 6.869101e-34 2
## DNAJC7 2.432184e-38 -0.4081540 0.074 0.353 8.902281e-34 2
## COPS3.1 2.850205e-38 -0.6918318 0.056 0.317 1.043232e-33 2
## UFM1.1 3.565517e-38 -0.5195842 0.077 0.356 1.305051e-33 2
## PNRC1.2 3.986821e-38 -0.4310637 0.356 0.785 1.459256e-33 2
## HNRNPF 4.061041e-38 -0.2619275 0.301 0.735 1.486422e-33 2
## MIF4GD.1 4.189330e-38 -1.3500949 0.027 0.262 1.533378e-33 2
## HCFC1R1.1 4.418434e-38 -2.4441887 0.020 0.246 1.617235e-33 2
## LYPLA1 5.104802e-38 -0.4874054 0.084 0.369 1.868460e-33 2
## SURF1.1 5.454286e-38 -1.3582791 0.035 0.274 1.996378e-33 2
## MRPL24 5.944031e-38 -1.3976504 0.029 0.264 2.175634e-33 2
## HSP90AA1.1 6.114148e-38 -0.5003475 0.417 0.845 2.237900e-33 2
## TSC22D4.1 6.542480e-38 -0.1972254 0.122 0.441 2.394678e-33 2
## WBP2.1 6.777836e-38 -0.7501219 0.065 0.332 2.480823e-33 2
## NDUFAF8.1 7.301296e-38 -1.0484868 0.039 0.284 2.672420e-33 2
## PCNA.2 7.978186e-38 -1.0300104 0.059 0.314 2.920176e-33 2
## GRSF1.1 1.009625e-37 -0.7531144 0.065 0.332 3.695431e-33 2
## UQCC2.2 1.078328e-37 -1.1321306 0.035 0.274 3.946897e-33 2
## MRPL38.1 1.102433e-37 -1.0840311 0.039 0.281 4.035126e-33 2
## RAB8A 1.109739e-37 -0.4546982 0.069 0.339 4.061868e-33 2
## RANGRF 1.209080e-37 -2.3515915 0.015 0.236 4.425475e-33 2
## DERL2.1 1.384709e-37 -1.4257382 0.033 0.271 5.068310e-33 2
## DNAJC15.1 1.545701e-37 -0.8374096 0.059 0.320 5.657573e-33 2
## GPS1 1.593844e-37 -1.1006028 0.038 0.278 5.833788e-33 2
## EIF2S2 1.638676e-37 -0.2631402 0.107 0.407 5.997883e-33 2
## EIF4E2.1 1.904570e-37 -0.4130492 0.065 0.331 6.971105e-33 2
## C18orf32.1 2.062100e-37 -1.1431179 0.033 0.270 7.547698e-33 2
## BABAM1.1 2.122807e-37 -0.6933454 0.057 0.315 7.769898e-33 2
## FAM32A 2.254599e-37 -0.9255308 0.044 0.290 8.252281e-33 2
## PLIN2.1 2.254952e-37 -0.3967430 0.059 0.319 8.253576e-33 2
## RTF2.1 2.748667e-37 -0.1367635 0.123 0.438 1.006067e-32 2
## JAGN1 3.329035e-37 -0.9296409 0.035 0.273 1.218493e-32 2
## CD8A.2 3.338902e-37 -0.9980014 0.206 0.509 1.222105e-32 2
## CRELD2.2 3.597649e-37 -0.9178610 0.059 0.316 1.316811e-32 2
## ANXA6.2 3.773248e-37 -0.1730565 0.260 0.681 1.381084e-32 2
## CTSZ 3.825238e-37 -1.6622740 0.029 0.259 1.400114e-32 2
## HTATIP2 4.543375e-37 -1.3764025 0.029 0.259 1.662966e-32 2
## ITGB7.2 4.652071e-37 -0.2512321 0.251 0.656 1.702751e-32 2
## VBP1 4.675572e-37 -0.6944225 0.038 0.278 1.711353e-32 2
## ATP5ME 5.521745e-37 -0.8937658 0.039 0.280 2.021069e-32 2
## DAXX.2 6.427324e-37 -0.4542659 0.086 0.367 2.352529e-32 2
## DRG1 6.619712e-37 -1.2610482 0.027 0.256 2.422947e-32 2
## NUDT22.1 9.164509e-37 -0.6273095 0.048 0.295 3.354394e-32 2
## E2F4 1.152567e-36 -0.1616674 0.063 0.325 4.218626e-32 2
## TPRKB 1.178725e-36 -1.8564124 0.017 0.234 4.314371e-32 2
## C4orf48.1 1.361484e-36 -0.5265525 0.065 0.326 4.983304e-32 2
## BAG1 1.438388e-36 -0.5725584 0.053 0.305 5.264787e-32 2
## GLO1.2 1.568529e-36 -0.2167095 0.141 0.461 5.741129e-32 2
## CDKN2A.2 1.587646e-36 -0.7859603 0.087 0.359 5.811102e-32 2
## MRPS18B 1.695232e-36 -0.8633357 0.045 0.288 6.204888e-32 2
## MTCH2 1.749845e-36 -0.2977995 0.072 0.341 6.404783e-32 2
## CSRP1.1 1.818400e-36 -1.1168561 0.035 0.268 6.655709e-32 2
## IFI35.2 1.883943e-36 -1.6731946 0.035 0.267 6.895607e-32 2
## RPL26L1 1.887010e-36 -1.5075318 0.017 0.233 6.906835e-32 2
## CTSB.2 1.928128e-36 -0.5244690 0.083 0.358 7.057336e-32 2
## MPST.2 2.015639e-36 -1.8113558 0.017 0.233 7.377644e-32 2
## RRP36 2.162830e-36 -0.6605722 0.045 0.289 7.916391e-32 2
## PPP6C 2.231508e-36 -1.0842996 0.047 0.290 8.167764e-32 2
## AC243960.1.2 2.514758e-36 -0.7200140 0.053 0.302 9.204518e-32 2
## HDDC2 2.516729e-36 -1.3630558 0.026 0.249 9.211732e-32 2
## HTATSF1.1 2.612301e-36 -0.2527089 0.156 0.488 9.561545e-32 2
## SLBP.2 2.825964e-36 -0.4684575 0.072 0.337 1.034359e-31 2
## NELFE 3.007456e-36 -1.0885517 0.024 0.246 1.100789e-31 2
## HDGF.1 3.061035e-36 -0.4971343 0.065 0.323 1.120400e-31 2
## RNASEH2B 3.112034e-36 -0.2335509 0.104 0.399 1.139067e-31 2
## EBPL 3.189703e-36 -0.8068126 0.038 0.273 1.167495e-31 2
## CDC123.1 3.303151e-36 -0.3521155 0.093 0.379 1.209019e-31 2
## PRPF6 3.383764e-36 -0.1320101 0.083 0.357 1.238525e-31 2
## MTLN 3.486224e-36 -1.5343798 0.021 0.240 1.276028e-31 2
## LMAN1 3.667722e-36 -0.2922120 0.131 0.442 1.342460e-31 2
## POP7.2 3.976231e-36 -2.0241007 0.012 0.223 1.455380e-31 2
## FRG1 4.241726e-36 -0.4066159 0.053 0.301 1.552557e-31 2
## RBBP7 5.262447e-36 -0.3753288 0.080 0.352 1.926161e-31 2
## ATP6AP2.1 7.106336e-36 -0.2601470 0.117 0.419 2.601061e-31 2
## HNRNPM 7.666072e-36 -0.3206384 0.159 0.493 2.805936e-31 2
## MAD2L2.2 7.735134e-36 -1.3973107 0.024 0.244 2.831214e-31 2
## ITGB1BP1.1 8.156114e-36 -0.5168934 0.072 0.336 2.985301e-31 2
## CAPZA2.1 1.012561e-35 -0.4162503 0.074 0.340 3.706177e-31 2
## MRPL12.1 1.077393e-35 -1.1310281 0.032 0.259 3.943473e-31 2
## CAPZB.2 1.117689e-35 -0.3193298 0.362 0.815 4.090964e-31 2
## HCLS1.2 1.120010e-35 -0.1604583 0.304 0.746 4.099459e-31 2
## GYG1.1 1.218100e-35 -0.7307576 0.063 0.317 4.458488e-31 2
## DHRS4.1 1.266096e-35 -1.4231283 0.026 0.246 4.634165e-31 2
## TPGS1.2 1.307478e-35 -0.4900703 0.056 0.304 4.785632e-31 2
## COMMD8.1 1.417392e-35 -1.3097700 0.027 0.249 5.187938e-31 2
## ZCRB1 1.549092e-35 -0.7081613 0.032 0.258 5.669985e-31 2
## STN1.1 1.622868e-35 -0.4944673 0.066 0.323 5.940020e-31 2
## YIPF3.1 1.685122e-35 -0.7075637 0.044 0.281 6.167883e-31 2
## TNFAIP8L2.1 1.694116e-35 -1.4381095 0.027 0.248 6.200805e-31 2
## POLR3K.1 1.751567e-35 -1.0976225 0.030 0.255 6.411087e-31 2
## PLEKHJ1.1 1.812974e-35 -0.6213335 0.044 0.281 6.635848e-31 2
## DOCK10.1 1.876491e-35 1.6635101 0.620 0.718 6.868333e-31 2
## TBC1D10C.2 1.880804e-35 0.1260572 0.241 0.649 6.884119e-31 2
## HNRNPUL1.1 2.194683e-35 -0.1321561 0.185 0.544 8.032978e-31 2
## SARS.1 2.489660e-35 -0.5415148 0.056 0.303 9.112654e-31 2
## EEF1E1 2.693590e-35 -1.5304412 0.021 0.236 9.859078e-31 2
## TSTA3 2.890871e-35 -0.8455682 0.042 0.277 1.058117e-30 2
## AP3S1 2.933376e-35 -0.7609942 0.057 0.305 1.073674e-30 2
## PSMD11.1 2.983550e-35 -0.6736230 0.057 0.306 1.092039e-30 2
## PARP1 2.997486e-35 -0.1437070 0.180 0.531 1.097140e-30 2
## DCPS.1 3.073367e-35 -1.1005554 0.039 0.270 1.124914e-30 2
## SMIM29.1 3.159783e-35 -1.0110066 0.032 0.257 1.156544e-30 2
## BID 3.655995e-35 -1.0474616 0.032 0.256 1.338167e-30 2
## MPLKIP 3.881838e-35 -0.9170896 0.033 0.259 1.420830e-30 2
## PEF1.1 4.145773e-35 -0.7054058 0.053 0.296 1.517436e-30 2
## PPCS.1 4.450301e-35 -0.6199248 0.057 0.305 1.628899e-30 2
## TNFRSF14.1 4.652360e-35 -0.1048765 0.155 0.481 1.702857e-30 2
## MRPL47 4.674747e-35 -0.8684213 0.035 0.262 1.711051e-30 2
## CBR1.2 4.734251e-35 -1.3697333 0.026 0.244 1.732831e-30 2
## UROD 5.681026e-35 -0.8383540 0.038 0.267 2.079369e-30 2
## CEBPB.1 6.039476e-35 -0.5650442 0.048 0.286 2.210569e-30 2
## TUBB4B 6.269781e-35 -0.1971285 0.226 0.601 2.294865e-30 2
## APOBEC3C.1 6.313457e-35 -1.0692863 0.038 0.266 2.310851e-30 2
## VMA21 7.886526e-35 -1.4870633 0.032 0.254 2.886626e-30 2
## GIMAP2.1 7.896236e-35 -0.6976159 0.060 0.307 2.890180e-30 2
## COMMD4.1 8.075238e-35 -1.1512255 0.035 0.260 2.955699e-30 2
## CNOT7.1 8.227316e-35 -1.0217793 0.050 0.289 3.011362e-30 2
## EXOSC1 8.514747e-35 -1.0555100 0.033 0.257 3.116568e-30 2
## DDA1.1 8.802521e-35 -1.0126781 0.032 0.253 3.221899e-30 2
## MED10.2 1.006941e-34 -0.3006016 0.081 0.348 3.685607e-30 2
## UBA2.1 1.032410e-34 -0.2519248 0.099 0.381 3.778827e-30 2
## CPNE3.1 1.090183e-34 -0.8221492 0.056 0.299 3.990288e-30 2
## PMPCB 1.125589e-34 -0.3290168 0.068 0.321 4.119881e-30 2
## UROS 1.154843e-34 -1.1267128 0.029 0.248 4.226955e-30 2
## RBM17 1.200286e-34 -0.1046053 0.120 0.419 4.393285e-30 2
## ZNF511 1.217657e-34 -1.5478237 0.023 0.236 4.456867e-30 2
## MRPS23 1.223115e-34 -0.9891641 0.032 0.253 4.476846e-30 2
## ABCE1 1.261341e-34 -0.8754783 0.042 0.273 4.616759e-30 2
## WBP1 1.278487e-34 -1.4169693 0.023 0.236 4.679518e-30 2
## HAUS1 1.287553e-34 -2.3053439 0.011 0.211 4.712703e-30 2
## MED28.1 1.352419e-34 -1.2170455 0.047 0.281 4.950123e-30 2
## MFF 1.410015e-34 -0.4789522 0.054 0.296 5.160939e-30 2
## CD96.1 1.488045e-34 1.2108942 0.686 0.812 5.446543e-30 2
## MLX.1 1.491409e-34 -0.6473470 0.044 0.277 5.458856e-30 2
## ANAPC15.1 1.675440e-34 -1.1058540 0.026 0.241 6.132446e-30 2
## PSMB5.1 1.677764e-34 -1.2296934 0.026 0.241 6.140951e-30 2
## MCRS1.1 1.742291e-34 -0.9971050 0.039 0.267 6.377135e-30 2
## CCNQ 1.797076e-34 -2.7415105 0.011 0.211 6.577656e-30 2
## MRPL22 2.059521e-34 -0.9979292 0.033 0.256 7.538260e-30 2
## USP1.1 2.091180e-34 -0.8759620 0.047 0.281 7.654138e-30 2
## MRPL3 2.138707e-34 -0.8468428 0.057 0.302 7.828097e-30 2
## CD5 2.228659e-34 -0.2761507 0.120 0.416 8.157338e-30 2
## ZNF22 2.284796e-34 -1.5119967 0.026 0.241 8.362809e-30 2
## TMEM18 2.406638e-34 -1.5943209 0.017 0.223 8.808777e-30 2
## ADIPOR1 2.529805e-34 -1.1710952 0.036 0.260 9.259591e-30 2
## TIPRL.1 2.674314e-34 -0.4439167 0.065 0.314 9.788525e-30 2
## TSR2 2.711676e-34 -1.5590726 0.023 0.234 9.925276e-30 2
## TMEM204.2 2.826733e-34 -0.9352920 0.050 0.285 1.034641e-29 2
## CSDE1.2 2.927389e-34 -0.1879656 0.209 0.582 1.071483e-29 2
## MRPL40 3.164717e-34 -1.2029427 0.026 0.240 1.158350e-29 2
## SHKBP1.1 3.187981e-34 -0.5881340 0.068 0.320 1.166865e-29 2
## PCNP.1 3.308916e-34 -0.1944680 0.117 0.412 1.211129e-29 2
## IKZF1 3.323467e-34 1.5482349 0.632 0.750 1.216456e-29 2
## ELOVL1.1 3.374557e-34 -0.4030245 0.047 0.281 1.235155e-29 2
## MPV17 3.518926e-34 -0.8460399 0.041 0.269 1.287997e-29 2
## TMX1.1 3.638412e-34 -0.5892595 0.063 0.311 1.331731e-29 2
## WDR61 3.675647e-34 -1.1461372 0.033 0.254 1.345360e-29 2
## LSM14A.1 3.872436e-34 -0.1372756 0.155 0.482 1.417389e-29 2
## AAMP 4.060863e-34 -0.9054060 0.041 0.269 1.486357e-29 2
## RTN3 4.818818e-34 -0.2074182 0.108 0.394 1.763784e-29 2
## HLA-DMA.1 4.856338e-34 -1.6978953 0.020 0.227 1.777517e-29 2
## TMEM14A 5.362359e-34 -2.7096270 0.011 0.208 1.962731e-29 2
## SH2D2A.1 5.477141e-34 -0.2422143 0.107 0.391 2.004743e-29 2
## DUSP23.1 6.028643e-34 -1.9017931 0.018 0.224 2.206604e-29 2
## PLAC8.2 6.827253e-34 -1.1187290 0.035 0.256 2.498911e-29 2
## TRAPPC4 7.100540e-34 -0.9090019 0.032 0.250 2.598940e-29 2
## RFXANK 7.128555e-34 -1.0332604 0.035 0.256 2.609194e-29 2
## DDRGK1 7.698630e-34 -0.6141122 0.068 0.316 2.817853e-29 2
## TERF2IP.1 8.971521e-34 0.2448948 0.186 0.542 3.283756e-29 2
## CYB561D2.2 9.861820e-34 -0.6846030 0.056 0.294 3.609623e-29 2
## SQOR.1 1.046299e-33 -0.2955347 0.066 0.314 3.829662e-29 2
## STXBP2.2 1.213234e-33 -0.1725039 0.093 0.364 4.440680e-29 2
## BCAS2 1.292220e-33 -0.7721986 0.056 0.292 4.729785e-29 2
## HM13 1.335098e-33 -0.1525234 0.150 0.468 4.886726e-29 2
## SSRP1.2 1.363774e-33 -0.5829168 0.056 0.294 4.991687e-29 2
## HLA-DRA.2 1.379605e-33 -0.9348934 0.074 0.322 5.049631e-29 2
## HDAC2.1 1.379719e-33 -1.0147321 0.042 0.268 5.050048e-29 2
## ADI1.2 1.613760e-33 -0.7958801 0.039 0.262 5.906684e-29 2
## TCP1.1 1.632303e-33 -0.3608651 0.096 0.366 5.974556e-29 2
## AHCY.2 2.057058e-33 -1.1258875 0.020 0.224 7.529242e-29 2
## POP4.1 2.086273e-33 -0.5536318 0.035 0.254 7.636176e-29 2
## MRPL58 2.167782e-33 -1.4844462 0.021 0.227 7.934514e-29 2
## THOC3.1 2.275580e-33 -0.3392365 0.068 0.315 8.329078e-29 2
## POLR2C 2.486437e-33 -1.1870758 0.023 0.230 9.100855e-29 2
## TFDP1.2 2.528935e-33 -0.6090861 0.062 0.304 9.256406e-29 2
## GBP4.1 2.591630e-33 -0.3657032 0.072 0.324 9.485886e-29 2
## SF3A3 2.605575e-33 -0.3224967 0.062 0.305 9.536926e-29 2
## HACD4.1 2.673659e-33 -0.5732423 0.059 0.299 9.786127e-29 2
## COMTD1 2.703278e-33 -1.0004983 0.035 0.252 9.894537e-29 2
## PNP.1 2.714828e-33 0.1740369 0.140 0.450 9.936812e-29 2
## SCNM1 3.064904e-33 -0.4150179 0.047 0.275 1.121816e-28 2
## TRIAP1.1 3.081182e-33 -1.3899354 0.024 0.232 1.127774e-28 2
## VAPA.1 3.151807e-33 -0.2833445 0.122 0.414 1.153624e-28 2
## EBP 3.324005e-33 -0.5533408 0.053 0.286 1.216652e-28 2
## TMEM183A 3.329125e-33 -0.8586997 0.036 0.255 1.218526e-28 2
## PPID 3.690655e-33 -1.2999754 0.023 0.229 1.350853e-28 2
## RUNX1.1 3.706515e-33 2.2796193 0.493 0.422 1.356659e-28 2
## YIF1B.1 3.811331e-33 -0.6771278 0.039 0.260 1.395023e-28 2
## THEMIS.1 4.065509e-33 1.8110514 0.603 0.685 1.488058e-28 2
## CAMLG 4.598279e-33 -0.6482245 0.060 0.299 1.683062e-28 2
## IMP4 4.826566e-33 -0.6954581 0.038 0.258 1.766620e-28 2
## SSR1 5.234989e-33 -0.2520974 0.102 0.376 1.916111e-28 2
## TMEM134 5.344800e-33 -0.8976017 0.032 0.245 1.956304e-28 2
## AIF1.1 5.504432e-33 -2.4806891 0.009 0.200 2.014732e-28 2
## EXOSC8.1 5.529469e-33 -0.3225859 0.054 0.289 2.023896e-28 2
## MRPS11 6.051600e-33 -1.0902818 0.029 0.239 2.215007e-28 2
## RGL4.2 6.190089e-33 -0.9388629 0.051 0.281 2.265696e-28 2
## RAD21 7.815137e-33 -0.2430809 0.173 0.500 2.860496e-28 2
## TFPT.1 7.899969e-33 -0.5813314 0.039 0.260 2.891547e-28 2
## GMPR2 8.463806e-33 -1.3133275 0.030 0.240 3.097922e-28 2
## SURF4.1 8.465881e-33 -0.2691870 0.116 0.401 3.098682e-28 2
## OXLD1 9.606212e-33 -0.5751194 0.044 0.268 3.516066e-28 2
## MZT1.1 1.032511e-32 -1.5256090 0.024 0.229 3.779197e-28 2
## POLDIP2 1.109793e-32 -1.3733864 0.020 0.221 4.062065e-28 2
## PGD.2 1.311922e-32 -1.2709145 0.023 0.226 4.801898e-28 2
## PRDX4.2 1.315388e-32 -1.6949399 0.015 0.211 4.814582e-28 2
## VPS72.1 1.322623e-32 -1.0438765 0.030 0.241 4.841064e-28 2
## SNAPIN 1.354239e-32 -0.9520238 0.029 0.238 4.956785e-28 2
## TINF2.1 1.414652e-32 -0.6298517 0.042 0.263 5.177910e-28 2
## TMEM126B 1.445553e-32 -1.2626785 0.021 0.223 5.291012e-28 2
## ZFAND2B.1 1.474789e-32 -0.9477897 0.036 0.252 5.398023e-28 2
## CELF2 1.565817e-32 1.8597350 0.573 0.615 5.731205e-28 2
## ATIC.1 1.613059e-32 -0.4020322 0.066 0.308 5.904119e-28 2
## RELA.1 1.643784e-32 -0.9131349 0.038 0.255 6.016579e-28 2
## AK2.1 1.734430e-32 -0.1886148 0.065 0.306 6.348360e-28 2
## NMI 1.776909e-32 -0.6206596 0.048 0.274 6.503841e-28 2
## MFSD10.2 1.884642e-32 -0.3677542 0.083 0.337 6.898167e-28 2
## ITGB2.2 1.888316e-32 -0.2316191 0.289 0.694 6.911613e-28 2
## EIF2S1 1.917250e-32 -0.6166473 0.047 0.272 7.017517e-28 2
## EIF2A 1.944942e-32 -0.2101147 0.087 0.346 7.118877e-28 2
## SIRPG.1 2.607273e-32 -0.5406720 0.065 0.302 9.543142e-28 2
## PTPN18 2.657893e-32 -0.3812256 0.048 0.273 9.728421e-28 2
## MRPS2 3.411311e-32 -1.6607370 0.021 0.220 1.248608e-27 2
## CDV3.1 3.437450e-32 0.1688494 0.203 0.562 1.258175e-27 2
## BCCIP 3.440370e-32 -0.4286355 0.059 0.293 1.259244e-27 2
## VPS51.2 3.628525e-32 -0.1385225 0.122 0.409 1.328113e-27 2
## LYSMD2 3.793450e-32 -2.1297416 0.009 0.197 1.388478e-27 2
## CCDC59 3.821507e-32 -0.9367387 0.038 0.252 1.398748e-27 2
## SDAD1 4.294344e-32 -0.6841512 0.060 0.293 1.571816e-27 2
## GNAS.1 4.299412e-32 -0.3050145 0.412 0.848 1.573671e-27 2
## SKAP1.2 4.624919e-32 1.4837062 0.641 0.790 1.692813e-27 2
## TMEM106C.2 4.670285e-32 -1.4341440 0.023 0.223 1.709418e-27 2
## WDR54 5.130113e-32 -1.0336071 0.045 0.266 1.877724e-27 2
## MRPL27.1 5.519685e-32 -1.0838435 0.035 0.246 2.020315e-27 2
## KPNB1.1 5.711017e-32 -0.1849530 0.138 0.436 2.090346e-27 2
## SFT2D1.1 5.772735e-32 -0.8868340 0.029 0.235 2.112936e-27 2
## DPM2.2 5.956502e-32 -1.0344403 0.021 0.220 2.180199e-27 2
## MRPL15 6.434557e-32 -2.1014586 0.015 0.208 2.355177e-27 2
## GIMAP5.2 6.487680e-32 -0.2457275 0.185 0.515 2.374621e-27 2
## CERS2 7.469743e-32 -0.6436561 0.042 0.259 2.734075e-27 2
## PSMD2 7.739642e-32 -0.4575095 0.083 0.334 2.832864e-27 2
## CLDND1.2 8.334483e-32 -0.1446113 0.214 0.566 3.050588e-27 2
## SNX5 8.591228e-32 -0.1420989 0.104 0.374 3.144561e-27 2
## RAB11A.1 9.582456e-32 0.1157614 0.194 0.546 3.507371e-27 2
## CTSS.2 9.615103e-32 -0.2523104 0.093 0.354 3.519320e-27 2
## ISCA2 9.631362e-32 -2.0007888 0.012 0.201 3.525271e-27 2
## ASF1A 1.003768e-31 -0.4816522 0.039 0.253 3.673990e-27 2
## GLIPR2.1 1.036667e-31 -0.2248090 0.087 0.343 3.794409e-27 2
## MRFAP1 1.066444e-31 -0.4448976 0.069 0.309 3.903397e-27 2
## BECN1.1 1.106610e-31 -0.4915208 0.062 0.295 4.050412e-27 2
## MRPS26.1 1.180421e-31 -0.4705800 0.053 0.279 4.320576e-27 2
## UPP1.2 1.207266e-31 0.1016634 0.128 0.417 4.418837e-27 2
## AASDHPPT.1 1.218290e-31 -0.9701734 0.042 0.258 4.459186e-27 2
## MRPL10.2 1.240023e-31 -0.5776182 0.051 0.276 4.538733e-27 2
## DNAJB11 1.315474e-31 -0.4895295 0.062 0.294 4.814900e-27 2
## DPP7 1.439842e-31 0.1771997 0.218 0.592 5.270108e-27 2
## BUD23 1.478283e-31 -0.7393895 0.038 0.249 5.410812e-27 2
## RNASEH2C.1 1.522388e-31 -0.7840090 0.026 0.227 5.572243e-27 2
## ZNF524 1.530436e-31 -1.5810006 0.026 0.225 5.601701e-27 2
## PDHB 1.553716e-31 -1.0797124 0.039 0.251 5.686911e-27 2
## STIP1.2 1.590126e-31 -0.5542617 0.083 0.333 5.820179e-27 2
## FARSA 1.629230e-31 -0.3723106 0.050 0.273 5.963308e-27 2
## CDIPT.1 1.836886e-31 -1.1550297 0.035 0.243 6.723368e-27 2
## MRPL32 1.846842e-31 -1.2292493 0.023 0.220 6.759811e-27 2
## FBXW5.2 1.855158e-31 0.2491689 0.125 0.412 6.790249e-27 2
## DOCK8.1 2.024367e-31 1.5676392 0.603 0.694 7.409589e-27 2
## TIMMDC1.1 2.039765e-31 -0.4793751 0.047 0.266 7.465949e-27 2
## NUDT16L1.1 2.387239e-31 -0.5604034 0.048 0.268 8.737773e-27 2
## HLA-DQB1.2 2.724730e-31 -0.3704381 0.114 0.383 9.973056e-27 2
## FNTA 2.859386e-31 -0.1861186 0.096 0.357 1.046593e-26 2
## PPP1R11.1 2.939236e-31 -0.3747700 0.071 0.311 1.075819e-26 2
## NCL 3.001463e-31 -0.2323721 0.359 0.788 1.098596e-26 2
## PDZD11 3.017309e-31 -1.7366484 0.015 0.204 1.104396e-26 2
## PPIE 3.176589e-31 -1.2650959 0.024 0.222 1.162695e-26 2
## MGAT1.2 3.205123e-31 -0.3759807 0.084 0.334 1.173139e-26 2
## NR1H2.1 3.225593e-31 -0.3820551 0.069 0.307 1.180632e-26 2
## SNRNP40 3.483820e-31 -0.3125912 0.065 0.299 1.275148e-26 2
## LCK.2 3.515042e-31 -0.1903693 0.368 0.808 1.286576e-26 2
## DDB1 3.774822e-31 -0.2754654 0.078 0.324 1.381660e-26 2
## DPYD 3.818791e-31 1.7725488 0.598 0.686 1.397754e-26 2
## ARL5A 4.204647e-31 -0.4760391 0.062 0.292 1.538985e-26 2
## MYCBP 4.272624e-31 -1.6009468 0.023 0.218 1.563866e-26 2
## MT-ATP8.2 4.824895e-31 0.2270025 0.663 0.702 1.766008e-26 2
## LAPTM4A.1 5.046122e-31 -0.7350105 0.047 0.264 1.846981e-26 2
## EML4.2 5.248843e-31 1.7578923 0.567 0.601 1.921182e-26 2
## TMEM167A 5.485902e-31 -0.5245745 0.063 0.294 2.007950e-26 2
## GMPS.1 5.521697e-31 -0.3540146 0.066 0.300 2.021052e-26 2
## ATP6AP1.1 5.951964e-31 -0.4896725 0.062 0.291 2.178538e-26 2
## PSMD3.1 6.525560e-31 -0.6172941 0.047 0.263 2.388486e-26 2
## TFAM 6.950739e-31 -1.0692593 0.035 0.240 2.544109e-26 2
## RALA 7.332067e-31 -0.6131285 0.062 0.290 2.683683e-26 2
## C20orf27.2 7.703866e-31 -1.4840574 0.020 0.211 2.819769e-26 2
## CCDC90B 7.965534e-31 -0.5243845 0.047 0.263 2.915545e-26 2
## PMF1.1 8.213778e-31 -0.3206705 0.057 0.282 3.006407e-26 2
## PDCL3.1 8.825106e-31 -1.3922585 0.018 0.208 3.230165e-26 2
## PCBD1.1 9.131650e-31 -1.9671978 0.015 0.202 3.342366e-26 2
## SYNC 9.852677e-31 -2.3359793 0.012 0.195 3.606277e-26 2
## RPP21 1.016757e-30 -0.5630958 0.042 0.253 3.721534e-26 2
## CYB5B 1.059223e-30 -0.3765431 0.065 0.295 3.876967e-26 2
## TCF7.2 1.063210e-30 -0.4190301 0.107 0.370 3.891560e-26 2
## BCKDHA 1.134076e-30 -1.2644561 0.020 0.209 4.150945e-26 2
## CHST11.1 1.177995e-30 2.3949504 0.499 0.456 4.311699e-26 2
## ISY1 1.206553e-30 -0.2930643 0.050 0.267 4.416224e-26 2
## SAMM50.1 1.297261e-30 -0.8180445 0.038 0.245 4.748234e-26 2
## TIMM17A 1.367356e-30 -0.5741703 0.051 0.270 5.004796e-26 2
## TRNAU1AP.1 1.435666e-30 -0.6085515 0.029 0.228 5.254825e-26 2
## UCHL5.1 1.586425e-30 -0.4649739 0.050 0.267 5.806634e-26 2
## ELAVL1.1 1.745142e-30 -0.3778568 0.068 0.299 6.387569e-26 2
## DNAJC4.1 1.768162e-30 -0.8033410 0.030 0.230 6.471827e-26 2
## ABT1.1 1.924600e-30 -1.2767175 0.038 0.243 7.044422e-26 2
## PBDC1.1 1.927937e-30 -0.9567192 0.036 0.241 7.056636e-26 2
## BLOC1S4.2 2.026830e-30 -0.3741152 0.041 0.249 7.418603e-26 2
## BCL7B 2.057528e-30 -0.1597006 0.053 0.273 7.530964e-26 2
## AIMP1 2.183477e-30 -0.2443718 0.063 0.291 7.991963e-26 2
## TMEM141.1 2.662729e-30 -0.8660098 0.032 0.232 9.746122e-26 2
## STMN1.2 2.700618e-30 -2.2236652 0.050 0.254 9.884801e-26 2
## STT3A 2.766405e-30 -0.5728218 0.068 0.297 1.012560e-25 2
## GNG10 2.863989e-30 -0.6772244 0.038 0.243 1.048277e-25 2
## MAD2L1.2 3.219081e-30 -2.4770386 0.005 0.178 1.178248e-25 2
## CISH.2 3.275969e-30 -0.2912949 0.313 0.689 1.199070e-25 2
## TMEM173.2 3.293865e-30 -0.2552916 0.125 0.402 1.205620e-25 2
## GTF2H5.1 3.502892e-30 -1.1994958 0.015 0.199 1.282129e-25 2
## SRGN.2 3.773620e-30 -0.2190980 0.358 0.777 1.381220e-25 2
## SNRNP25.2 3.929040e-30 -1.8065982 0.015 0.198 1.438107e-25 2
## MTFP1.1 4.160686e-30 -0.4656660 0.035 0.237 1.522894e-25 2
## H2AFX.2 4.240679e-30 -0.4963284 0.074 0.306 1.552173e-25 2
## PABPC1.2 4.665284e-30 -0.3887605 0.499 0.886 1.707587e-25 2
## NCOA4.1 4.864246e-30 -0.3154506 0.105 0.365 1.780411e-25 2
## GPAA1.2 4.876971e-30 -0.5316045 0.054 0.273 1.785069e-25 2
## GRK6.1 5.089838e-30 0.1754331 0.129 0.415 1.862983e-25 2
## SNRPN 5.809118e-30 -1.4317052 0.023 0.212 2.126253e-25 2
## KLHDC3 6.692870e-30 -0.9604895 0.030 0.227 2.449724e-25 2
## NDUFAF4 6.743774e-30 -1.2877558 0.015 0.197 2.468356e-25 2
## UBE2V2 7.280914e-30 -0.2350586 0.060 0.284 2.664960e-25 2
## PRPF31 7.583852e-30 -0.6015199 0.041 0.247 2.775842e-25 2
## HNRNPAB.2 7.889942e-30 -0.1321076 0.116 0.380 2.887877e-25 2
## INO80E.1 8.155441e-30 -0.1622378 0.065 0.292 2.985055e-25 2
## PIGT.1 9.010864e-30 -0.6930742 0.039 0.244 3.298156e-25 2
## MTHFS.1 9.736989e-30 -1.1883976 0.023 0.211 3.563933e-25 2
## PSMC1 1.009739e-29 -0.6612659 0.050 0.262 3.695848e-25 2
## SLC25A1.1 1.026429e-29 -1.4710887 0.020 0.205 3.756935e-25 2
## MPC1 1.040482e-29 -1.2036886 0.027 0.220 3.808371e-25 2
## GLOD4 1.048829e-29 -0.8617589 0.038 0.240 3.838923e-25 2
## BAD.1 1.066102e-29 -0.5073585 0.039 0.243 3.902146e-25 2
## LCMT1 1.072667e-29 -1.0023285 0.035 0.235 3.926175e-25 2
## LST1.2 1.185109e-29 -0.9077290 0.021 0.208 4.337735e-25 2
## DHPS 1.382196e-29 -0.4054839 0.054 0.270 5.059114e-25 2
## ATOX1.2 1.545351e-29 -0.5893289 0.035 0.233 5.656293e-25 2
## MRPL37.1 1.571816e-29 -0.2521794 0.050 0.262 5.753162e-25 2
## ZFAS1.2 1.666331e-29 0.3885889 0.269 0.673 6.099104e-25 2
## PAGR1.1 1.865883e-29 -0.4273048 0.047 0.256 6.829504e-25 2
## MAPKAPK5-AS1.1 1.896751e-29 -0.7610631 0.039 0.241 6.942489e-25 2
## MAPK1IP1L 1.939641e-29 -0.1088900 0.116 0.384 7.099476e-25 2
## CIRBP.2 2.154455e-29 0.1373274 0.271 0.674 7.885736e-25 2
## SRPRB 2.312750e-29 -1.0229139 0.044 0.249 8.465128e-25 2
## ACAT2.2 2.379573e-29 -1.2521228 0.030 0.224 8.709715e-25 2
## CDK2AP1.2 2.439828e-29 -1.7400491 0.017 0.197 8.930257e-25 2
## TMEM243.1 2.446903e-29 -0.2100673 0.063 0.286 8.956156e-25 2
## CMTM7.1 2.514081e-29 -0.2497650 0.066 0.292 9.202041e-25 2
## ZNRD2 2.637578e-29 -0.5401001 0.030 0.224 9.654064e-25 2
## ALDH9A1.1 2.640065e-29 -0.6816532 0.044 0.249 9.663166e-25 2
## PTTG1.1 2.690089e-29 -2.2630358 0.017 0.196 9.846263e-25 2
## MOB4.1 2.917020e-29 -0.4746136 0.056 0.271 1.067688e-24 2
## CAPN1.1 2.945893e-29 -0.3101072 0.090 0.334 1.078256e-24 2
## DKC1 3.137367e-29 -1.3900337 0.021 0.206 1.148339e-24 2
## TXNDC9 3.222305e-29 -0.4974043 0.036 0.234 1.179428e-24 2
## C1orf122.1 3.283800e-29 -0.7433723 0.035 0.232 1.201937e-24 2
## GTPBP6.1 3.415697e-29 -0.2973964 0.047 0.254 1.250214e-24 2
## MED30.1 3.438868e-29 -0.9668123 0.027 0.217 1.258695e-24 2
## SEPTIN2.1 3.524274e-29 -0.1099267 0.138 0.421 1.289955e-24 2
## RPL7L1 3.684694e-29 -0.7598996 0.051 0.261 1.348672e-24 2
## ANAPC13 3.957645e-29 -1.8441475 0.014 0.191 1.448577e-24 2
## RAB24.1 4.266425e-29 -0.6954750 0.044 0.247 1.561597e-24 2
## PNRC2 4.339016e-29 -0.1334036 0.119 0.388 1.588167e-24 2
## SNHG7.1 4.411084e-29 0.2367821 0.113 0.378 1.614545e-24 2
## NTMT1 4.523515e-29 -1.3152459 0.021 0.205 1.655697e-24 2
## YTHDF2 4.797107e-29 -0.1751073 0.089 0.333 1.755837e-24 2
## EIF3A.1 4.967108e-29 0.2051905 0.223 0.584 1.818061e-24 2
## MRPS14 5.066025e-29 -1.0957654 0.026 0.214 1.854266e-24 2
## KARS 5.667425e-29 -0.6221647 0.042 0.244 2.074391e-24 2
## HMOX2.2 7.370884e-29 0.1315449 0.164 0.472 2.697891e-24 2
## RPS10 8.468525e-29 -1.8576098 0.015 0.192 3.099650e-24 2
## POLD2.2 8.472453e-29 -1.2354796 0.024 0.209 3.101087e-24 2
## GPR108.1 9.093230e-29 -0.1523746 0.069 0.295 3.328304e-24 2
## DTYMK.2 9.632309e-29 -1.4692204 0.021 0.203 3.525618e-24 2
## CALCOCO2.2 1.031367e-28 -0.2319056 0.101 0.350 3.775009e-24 2
## CYCS.1 1.125496e-28 0.1653198 0.149 0.445 4.119541e-24 2
## ANKRD12 1.292448e-28 1.6075922 0.598 0.689 4.730619e-24 2
## PSMC6 1.316902e-28 -0.1747918 0.074 0.302 4.820126e-24 2
## VTI1B.2 1.335482e-28 -0.4849548 0.048 0.254 4.888132e-24 2
## SIGIRR 1.340453e-28 0.3766481 0.153 0.453 4.906326e-24 2
## RFLNB.2 1.348186e-28 -0.4577045 0.057 0.271 4.934632e-24 2
## CTBP1 1.685495e-28 -0.1261497 0.074 0.300 6.169248e-24 2
## PSMG1.2 1.717714e-28 -0.7849322 0.029 0.217 6.287176e-24 2
## CAPZA1.1 1.759649e-28 0.1390804 0.192 0.522 6.440669e-24 2
## EBNA1BP2.1 1.920017e-28 -1.3704766 0.023 0.204 7.027648e-24 2
## MVP.1 1.923776e-28 0.1459246 0.134 0.413 7.041405e-24 2
## BTF3L4 1.965337e-28 -0.4340377 0.056 0.268 7.193528e-24 2
## SEPHS2.2 2.055236e-28 0.1825841 0.093 0.335 7.522576e-24 2
## EIF1AX.1 2.059712e-28 -0.1683873 0.062 0.278 7.538957e-24 2
## HAGH.1 2.087530e-28 -0.2134637 0.059 0.272 7.640776e-24 2
## ZFAND1.1 2.213317e-28 -1.1599291 0.027 0.213 8.101182e-24 2
## PHF11 2.337591e-28 -0.4418885 0.062 0.277 8.556052e-24 2
## ABHD14A 2.385605e-28 -1.4960800 0.017 0.192 8.731790e-24 2
## RBBP4 2.652109e-28 0.1962002 0.155 0.450 9.707248e-24 2
## BRMS1 2.802984e-28 -0.6961126 0.042 0.241 1.025948e-23 2
## CDC25B.2 2.805150e-28 -0.3287295 0.056 0.266 1.026741e-23 2
## PYM1 3.115982e-28 -0.7449246 0.035 0.226 1.140512e-23 2
## PDIA4 3.190175e-28 -0.4617239 0.078 0.306 1.167668e-23 2
## ZNF581.1 3.204643e-28 -1.5264952 0.018 0.195 1.172964e-23 2
## EAPP 3.240663e-28 -0.3917170 0.059 0.271 1.186147e-23 2
## SIGMAR1 3.456539e-28 -0.7963507 0.039 0.234 1.265162e-23 2
## UBC.2 3.858153e-28 -0.3301233 0.594 0.903 1.412161e-23 2
## RUVBL1 3.865435e-28 -0.8812571 0.029 0.214 1.414826e-23 2
## CGGBP1 4.058873e-28 -0.1081365 0.107 0.358 1.485629e-23 2
## LAIR1.1 4.221839e-28 -0.2870028 0.068 0.286 1.545278e-23 2
## COPS8 4.473849e-28 -0.5463148 0.044 0.242 1.637518e-23 2
## PAICS.2 4.512196e-28 -0.7101062 0.033 0.223 1.651554e-23 2
## PAPOLA 4.534852e-28 0.1639213 0.147 0.437 1.659847e-23 2
## GID8.1 4.817909e-28 -0.8646705 0.033 0.223 1.763451e-23 2
## NSMCE3 4.964995e-28 -0.4664823 0.053 0.259 1.817288e-23 2
## PLD3.2 5.094493e-28 -0.2054127 0.083 0.315 1.864686e-23 2
## GSK3A.1 5.244846e-28 -0.6210121 0.039 0.233 1.919719e-23 2
## NOB1 5.534861e-28 -0.7250048 0.029 0.214 2.025870e-23 2
## TSEN15.1 6.068937e-28 -0.5279226 0.056 0.265 2.221352e-23 2
## PRPS1 6.340983e-28 -1.4121981 0.020 0.196 2.320927e-23 2
## LGALS3BP.2 6.952351e-28 -0.8106484 0.041 0.237 2.544700e-23 2
## NXT1.2 7.197542e-28 -1.1902715 0.033 0.221 2.634444e-23 2
## C1D 7.591533e-28 -0.7961236 0.038 0.230 2.778653e-23 2
## DECR1.1 8.080216e-28 0.1754827 0.125 0.392 2.957521e-23 2
## UTRN.1 8.265662e-28 1.9096813 0.532 0.545 3.025398e-23 2
## QARS 8.619742e-28 -0.3985151 0.066 0.282 3.154998e-23 2
## VPS25.1 9.735105e-28 -0.6989994 0.033 0.221 3.563243e-23 2
## MT-ND3.2 9.864343e-28 0.4120816 0.663 0.752 3.610547e-23 2
## S1PR1.2 1.016554e-27 -0.3483816 0.062 0.273 3.720791e-23 2
## NIT2.1 1.017727e-27 -1.0317403 0.032 0.217 3.725084e-23 2
## PRPF38A 1.081321e-27 -0.8215909 0.050 0.251 3.957852e-23 2
## IDH3B 1.142982e-27 -0.4028345 0.039 0.232 4.183544e-23 2
## WDR77.2 1.203786e-27 -1.0946362 0.029 0.212 4.406098e-23 2
## USP39.1 1.325594e-27 -0.6494737 0.044 0.240 4.851938e-23 2
## PKN1.1 1.347808e-27 0.2027926 0.144 0.426 4.933248e-23 2
## CMAS 1.461243e-27 -1.3369698 0.018 0.191 5.348441e-23 2
## NCF4.1 1.511740e-27 -0.2094204 0.065 0.279 5.533270e-23 2
## LRP10.2 1.533089e-27 -0.4151549 0.080 0.306 5.611411e-23 2
## ORMDL3.1 1.670125e-27 -0.3601457 0.057 0.265 6.112990e-23 2
## KRR1 1.751728e-27 -0.4578780 0.066 0.281 6.411676e-23 2
## ITM2C.2 1.784151e-27 -0.6915177 0.053 0.255 6.530349e-23 2
## C15orf61 1.857021e-27 -1.9312062 0.014 0.182 6.797067e-23 2
## MPDU1.1 1.859259e-27 -0.9598353 0.032 0.217 6.805260e-23 2
## ANAPC5 1.977489e-27 0.1982469 0.155 0.447 7.238007e-23 2
## PLGRKT 2.130796e-27 -0.6538292 0.032 0.217 7.799139e-23 2
## HLA-DRB5.1 2.223184e-27 -0.7440207 0.072 0.287 8.137299e-23 2
## MITD1.1 2.572628e-27 -0.2627262 0.042 0.236 9.416332e-23 2
## AMZ2 2.665098e-27 -0.4014775 0.060 0.269 9.754791e-23 2
## ACAA1 3.336532e-27 -0.1617131 0.066 0.281 1.221238e-22 2
## G6PD.1 3.382977e-27 -1.0306695 0.036 0.223 1.238237e-22 2
## WDR18 3.475540e-27 -0.5628020 0.035 0.221 1.272117e-22 2
## SMARCD2 3.628517e-27 -0.5802463 0.051 0.252 1.328110e-22 2
## DCTD 3.863297e-27 -0.9010366 0.026 0.204 1.414044e-22 2
## GGCT.2 3.864859e-27 -0.5280169 0.026 0.204 1.414616e-22 2
## DARS 4.209389e-27 0.2024649 0.128 0.392 1.540721e-22 2
## MMP24OS.2 4.469397e-27 -0.1077144 0.084 0.312 1.635889e-22 2
## CCDC115.1 4.493503e-27 -1.5917990 0.026 0.203 1.644712e-22 2
## RRM1.2 4.587657e-27 -0.6326053 0.050 0.248 1.679174e-22 2
## SNAP29 5.381322e-27 -0.3274628 0.042 0.235 1.969671e-22 2
## MGMT 5.396789e-27 0.2044899 0.110 0.359 1.975333e-22 2
## MCRIP1.1 5.537793e-27 -1.4925072 0.015 0.182 2.026943e-22 2
## RAMAC 6.213609e-27 -1.4881397 0.017 0.185 2.274305e-22 2
## SLC35D2.2 6.247054e-27 -0.4970672 0.048 0.243 2.286547e-22 2
## HNRNPH2.1 6.400796e-27 -0.2215543 0.063 0.273 2.342819e-22 2
## HDDC3 6.460302e-27 -0.4937551 0.029 0.208 2.364600e-22 2
## KIF22.2 6.768099e-27 -0.4323807 0.063 0.271 2.477260e-22 2
## RAB18.2 6.827846e-27 -0.1066245 0.075 0.296 2.499128e-22 2
## B3GAT3 6.839205e-27 -0.2043174 0.063 0.273 2.503286e-22 2
## MAGED2.1 6.867517e-27 -1.5540136 0.026 0.202 2.513649e-22 2
## TIMM50.1 6.989549e-27 -0.7186593 0.021 0.194 2.558315e-22 2
## CYBC1.1 7.556344e-27 0.1770335 0.119 0.375 2.765773e-22 2
## NIPSNAP1 7.775589e-27 -1.1533634 0.029 0.208 2.846021e-22 2
## LTA4H 8.014231e-27 -0.5770525 0.042 0.233 2.933369e-22 2
## VPS35 8.770902e-27 -0.2129927 0.069 0.284 3.210326e-22 2
## MED11.1 9.517843e-27 -1.3851500 0.024 0.198 3.483721e-22 2
## GTF2F1 1.008251e-26 0.1717265 0.081 0.306 3.690402e-22 2
## LRRC59.1 1.023167e-26 -0.6152312 0.044 0.236 3.744997e-22 2
## FAM50A.1 1.029762e-26 0.3891432 0.102 0.346 3.769134e-22 2
## PITHD1 1.031583e-26 -0.4953695 0.047 0.241 3.775800e-22 2
## MID1IP1.1 1.098004e-26 -0.6650915 0.045 0.237 4.018914e-22 2
## TUSC2.1 1.120054e-26 -0.7580547 0.029 0.207 4.099621e-22 2
## PPIG.2 1.166878e-26 0.1026925 0.147 0.425 4.271009e-22 2
## APOBEC3H.2 1.182116e-26 -0.7896601 0.042 0.231 4.326781e-22 2
## MRPL48 1.308925e-26 -0.7781815 0.027 0.203 4.790926e-22 2
## RNF113A.1 1.318332e-26 -0.4608988 0.044 0.234 4.825359e-22 2
## ZNF593.1 1.455775e-26 -0.7994758 0.023 0.195 5.328429e-22 2
## ATXN1.1 1.463835e-26 2.2634333 0.481 0.446 5.357927e-22 2
## COMMD5.1 1.515019e-26 -1.2692563 0.026 0.200 5.545274e-22 2
## CFAP298 1.574839e-26 -0.4712707 0.047 0.240 5.764225e-22 2
## TSN 1.822989e-26 -1.0300266 0.032 0.211 6.672506e-22 2
## GOT2.1 1.852427e-26 -0.7880453 0.029 0.206 6.780253e-22 2
## PET100.1 2.052523e-26 -0.2464314 0.038 0.223 7.512645e-22 2
## CD59 2.083750e-26 -1.2890463 0.024 0.196 7.626941e-22 2
## FAM136A 2.136468e-26 -0.2137672 0.054 0.254 7.819901e-22 2
## CCDC28A.1 2.136776e-26 -1.7782228 0.015 0.179 7.821029e-22 2
## NTAN1.1 2.159784e-26 -0.8861796 0.029 0.205 7.905242e-22 2
## PDE6D.1 2.291038e-26 -0.3267098 0.041 0.228 8.385656e-22 2
## CCDC28B.1 2.552955e-26 -1.6158860 0.009 0.167 9.344327e-22 2
## MYC.2 2.655112e-26 -0.6198376 0.065 0.269 9.718240e-22 2
## UBTF.1 2.703549e-26 0.2316712 0.113 0.358 9.895530e-22 2
## STK16 2.844626e-26 -1.7622163 0.018 0.184 1.041190e-21 2
## ANXA4.1 2.969346e-26 -0.3433651 0.062 0.267 1.086840e-21 2
## NCBP2AS2.1 3.523895e-26 -0.3175792 0.039 0.224 1.289816e-21 2
## GNPDA1 3.534576e-26 -0.6290511 0.042 0.230 1.293725e-21 2
## SF1.1 3.594652e-26 0.1912301 0.235 0.586 1.315715e-21 2
## FDX1 3.616271e-26 -0.5056066 0.045 0.235 1.323627e-21 2
## NOP56 3.720454e-26 0.1355060 0.087 0.311 1.361760e-21 2
## SLC3A2 3.751749e-26 0.1785999 0.134 0.398 1.373215e-21 2
## FEN1.2 3.781710e-26 -1.9048610 0.015 0.177 1.384181e-21 2
## UBLCP1.1 3.919112e-26 -0.6855832 0.047 0.238 1.434473e-21 2
## BAK1 3.939440e-26 -0.7366269 0.039 0.224 1.441914e-21 2
## TMEM248.1 4.031984e-26 -0.1956569 0.074 0.287 1.475787e-21 2
## ASAH1 4.077949e-26 -0.5677005 0.042 0.229 1.492611e-21 2
## MALSU1.1 4.142392e-26 -0.7642571 0.026 0.198 1.516199e-21 2
## SSBP4.2 4.333001e-26 0.3454239 0.220 0.558 1.585965e-21 2
## PFKL 4.853887e-26 0.3054417 0.087 0.313 1.776620e-21 2
## MCM7.2 4.889678e-26 -0.9947944 0.047 0.233 1.789720e-21 2
## SUPT16H.1 4.982754e-26 -0.1325821 0.116 0.364 1.823788e-21 2
## RAB7A.1 5.105483e-26 0.1599390 0.211 0.540 1.868709e-21 2
## MED8 5.232592e-26 -2.4633305 0.012 0.171 1.915233e-21 2
## TIMM23.1 5.261747e-26 -0.5729574 0.048 0.240 1.925905e-21 2
## NSFL1C 5.344385e-26 -0.6965637 0.036 0.218 1.956152e-21 2
## LINC02446.2 5.657000e-26 -1.3396397 0.045 0.229 2.070575e-21 2
## HSPA4 5.760525e-26 -0.1256009 0.072 0.284 2.108467e-21 2
## TMEM223 5.858167e-26 -0.4548316 0.038 0.220 2.144206e-21 2
## USB1 6.508630e-26 -0.4026645 0.035 0.214 2.382289e-21 2
## CDC42SE2.1 6.517099e-26 1.5788644 0.592 0.697 2.385389e-21 2
## ACTR10.1 6.915583e-26 -0.2912303 0.042 0.229 2.531242e-21 2
## RPS6KB2 7.020800e-26 -0.4169909 0.039 0.222 2.569753e-21 2
## SUGT1 7.063227e-26 0.1941136 0.078 0.296 2.585282e-21 2
## DUS1L 7.070864e-26 -0.1000473 0.078 0.294 2.588078e-21 2
## CEP57 7.201239e-26 0.1365311 0.122 0.373 2.635797e-21 2
## RPIA.1 7.384366e-26 -0.4369262 0.054 0.249 2.702826e-21 2
## DNAJC9.2 7.608732e-26 -0.5424164 0.044 0.231 2.784948e-21 2
## DAP3 8.327187e-26 0.2352084 0.132 0.395 3.047917e-21 2
## EZR.2 8.819450e-26 0.1633582 0.253 0.609 3.228095e-21 2
## DDX39B.1 8.853213e-26 0.2399178 0.215 0.544 3.240453e-21 2
## COPB2 9.147593e-26 0.2520366 0.117 0.365 3.348202e-21 2
## ATF4 9.231772e-26 -0.1684851 0.099 0.330 3.379013e-21 2
## TRAT1.1 9.286312e-26 -0.5860395 0.077 0.288 3.398976e-21 2
## THOC6.1 9.613055e-26 -0.8891188 0.020 0.184 3.518571e-21 2
## OLA1.1 9.833250e-26 0.2244157 0.140 0.406 3.599166e-21 2
## GALK1.1 9.953750e-26 -0.7355832 0.027 0.199 3.643271e-21 2
## SMC4.2 1.027326e-25 -0.4875761 0.087 0.306 3.760219e-21 2
## NOL12.1 1.034121e-25 -0.5708077 0.036 0.216 3.785090e-21 2
## HADH.2 1.156005e-25 -0.5361197 0.027 0.199 4.231209e-21 2
## RILPL2 1.180910e-25 -0.1209582 0.060 0.260 4.322368e-21 2
## FYTTD1 1.257889e-25 -0.5469054 0.056 0.252 4.604125e-21 2
## KEAP1.1 1.281134e-25 -0.3124737 0.054 0.249 4.689207e-21 2
## UTP3 1.340120e-25 -1.6086896 0.018 0.180 4.905106e-21 2
## SMNDC1 1.400173e-25 -0.3121282 0.074 0.284 5.124914e-21 2
## CRBN.2 1.480288e-25 -0.1789253 0.095 0.321 5.418150e-21 2
## MRPL55 1.516433e-25 -0.3885746 0.035 0.212 5.550449e-21 2
## SLC35B2 1.560683e-25 -1.5783769 0.023 0.189 5.712413e-21 2
## UBE2J2.1 1.614088e-25 -0.5909161 0.045 0.231 5.907884e-21 2
## MCM3.2 1.851168e-25 -0.5779794 0.072 0.277 6.775645e-21 2
## RPP25L 2.004078e-25 -2.2155038 0.011 0.165 7.335325e-21 2
## MOB2.2 2.017579e-25 -0.4909787 0.042 0.225 7.384744e-21 2
## DDIT4.2 2.048026e-25 -0.1045775 0.162 0.433 7.496183e-21 2
## TXNDC15 2.063846e-25 -0.1964972 0.062 0.260 7.554089e-21 2
## TOR1A 2.314740e-25 -0.8502551 0.032 0.206 8.472412e-21 2
## AK6 2.433217e-25 -2.0908911 0.009 0.161 8.906059e-21 2
## NGRN 2.452692e-25 -0.7111587 0.033 0.208 8.977344e-21 2
## SPN.2 2.470801e-25 0.1459494 0.168 0.455 9.043627e-21 2
## CALHM2.1 2.501944e-25 -0.6062892 0.057 0.250 9.157615e-21 2
## MTIF3.1 2.508992e-25 -0.3406178 0.057 0.253 9.183414e-21 2
## CIAPIN1.1 2.618554e-25 -1.4635795 0.020 0.182 9.584433e-21 2
## SMS.1 2.633374e-25 -0.8336840 0.026 0.194 9.638674e-21 2
## GBP1.2 2.660648e-25 -0.3212211 0.072 0.277 9.738503e-21 2
## CZIB 2.677624e-25 -0.5563163 0.032 0.205 9.800638e-21 2
## CCDC25 3.014373e-25 -0.6335705 0.038 0.215 1.103321e-20 2
## MCRIP2 3.051724e-25 -2.7046082 0.008 0.158 1.116992e-20 2
## C7orf50.1 3.057223e-25 -0.1162652 0.056 0.249 1.119005e-20 2
## ERG28 3.080016e-25 -0.6692538 0.032 0.205 1.127347e-20 2
## SVIP 3.085486e-25 -1.3277440 0.020 0.181 1.129349e-20 2
## MBOAT7.2 3.167510e-25 -0.3080674 0.054 0.245 1.159372e-20 2
## COMMD9 3.176106e-25 -1.5036403 0.018 0.179 1.162518e-20 2
## GLMP.1 3.302847e-25 -1.8134912 0.017 0.176 1.208908e-20 2
## MRPS10 3.543863e-25 -0.2158829 0.041 0.222 1.297125e-20 2
## TOX4.1 3.577071e-25 0.3951309 0.092 0.315 1.309279e-20 2
## COQ4 3.768109e-25 -0.3763554 0.042 0.224 1.379203e-20 2
## U2AF1L4 3.863309e-25 -0.1454362 0.050 0.238 1.414048e-20 2
## ARL14EP.1 4.237525e-25 -0.2675705 0.072 0.278 1.551019e-20 2
## CS.1 4.294710e-25 -0.1563268 0.065 0.265 1.571950e-20 2
## RNPEPL1.2 4.349241e-25 0.1312633 0.147 0.415 1.591909e-20 2
## FTSJ1 4.530081e-25 -0.4136499 0.038 0.215 1.658100e-20 2
## RAE1 4.634661e-25 -0.9664830 0.033 0.207 1.696379e-20 2
## NOP16 5.041315e-25 -0.9702037 0.023 0.187 1.845222e-20 2
## INTS11 5.253496e-25 -0.1090689 0.075 0.283 1.922885e-20 2
## RWDD3 5.618444e-25 -1.5483160 0.017 0.174 2.056463e-20 2
## MRPL2 5.783999e-25 -1.8814848 0.015 0.172 2.117059e-20 2
## ALG3 5.827111e-25 -0.8085819 0.030 0.200 2.132839e-20 2
## ZNF24 5.965295e-25 -0.3078587 0.066 0.267 2.183417e-20 2
## GNPTG 6.105524e-25 -0.4534783 0.044 0.225 2.234744e-20 2
## TIAL1 6.568841e-25 0.1490061 0.116 0.358 2.404327e-20 2
## ADD3.2 6.874357e-25 0.3544421 0.191 0.494 2.516152e-20 2
## CD55.1 7.166999e-25 0.2699297 0.195 0.502 2.623265e-20 2
## GOLT1B 7.449959e-25 -0.7952471 0.042 0.222 2.726834e-20 2
## C11orf1 7.954352e-25 -2.7012901 0.005 0.150 2.911452e-20 2
## PAIP1 8.012294e-25 -0.6846873 0.035 0.208 2.932660e-20 2
## SNAPC5 8.514899e-25 -1.2713683 0.015 0.171 3.116623e-20 2
## EI24 8.597624e-25 -0.8595535 0.026 0.191 3.146902e-20 2
## PPM1M.1 8.697451e-25 -0.6752100 0.045 0.227 3.183441e-20 2
## C16orf54.2 8.735649e-25 -0.8380408 0.038 0.213 3.197422e-20 2
## RAB4A 8.820055e-25 -0.6378876 0.038 0.213 3.228317e-20 2
## SPTSSA 9.029653e-25 -2.1697852 0.012 0.164 3.305033e-20 2
## RTCB.1 9.997681e-25 -0.6357345 0.038 0.212 3.659351e-20 2
## FBXL15 1.000190e-24 -1.2481993 0.018 0.176 3.660895e-20 2
## EIF4EBP1.2 1.040634e-24 -1.6879723 0.009 0.158 3.808928e-20 2
## SKA2 1.061795e-24 -0.8229586 0.033 0.204 3.886384e-20 2
## EXOSC5.1 1.160135e-24 -0.5952763 0.035 0.207 4.246325e-20 2
## DNAAF2 1.174128e-24 -0.4885968 0.035 0.207 4.297544e-20 2
## SLC10A3.1 1.211513e-24 -1.4746684 0.021 0.181 4.434381e-20 2
## TRAF3IP3.2 1.268612e-24 0.1333967 0.317 0.721 4.643374e-20 2
## CAT 1.309405e-24 -0.2650029 0.056 0.246 4.792685e-20 2
## MRPS25.2 1.397704e-24 -0.3827430 0.036 0.210 5.115875e-20 2
## FH.2 1.405891e-24 -0.9031720 0.023 0.184 5.145842e-20 2
## NDUFS4 1.477543e-24 -0.4045616 0.065 0.262 5.408103e-20 2
## HERPUD1.1 1.782913e-24 0.2763794 0.131 0.381 6.525818e-20 2
## HSD17B11.2 1.800990e-24 -0.4342718 0.036 0.209 6.591984e-20 2
## RNF220.1 1.801654e-24 -0.1482405 0.054 0.243 6.594415e-20 2
## ITGAE 1.880372e-24 -0.6026385 0.035 0.206 6.882537e-20 2
## SH2D1A.1 1.905781e-24 -0.3405113 0.060 0.253 6.975539e-20 2
## UBE2T.2 1.959184e-24 -1.5232804 0.017 0.171 7.171006e-20 2
## ABI3.1 1.994533e-24 -0.2053949 0.080 0.287 7.300388e-20 2
## ADAM19.1 2.097534e-24 2.3702355 0.409 0.326 7.677395e-20 2
## DNASE2.1 2.162679e-24 -1.3398024 0.020 0.177 7.915836e-20 2
## SAC3D1.2 2.301875e-24 -2.0252330 0.008 0.153 8.425324e-20 2
## TIMM9.1 2.310117e-24 -0.6610239 0.026 0.189 8.455490e-20 2
## EIF2B1 2.584187e-24 -0.4987872 0.044 0.222 9.458642e-20 2
## HP1BP3.1 2.586615e-24 0.1206862 0.263 0.615 9.467528e-20 2
## AKIP1.1 2.618598e-24 -1.5506009 0.014 0.164 9.584593e-20 2
## TMEM205 2.653030e-24 -2.2058298 0.014 0.165 9.710620e-20 2
## AKIRIN1.1 2.673634e-24 -0.1966005 0.077 0.281 9.786036e-20 2
## DNPEP.1 2.730609e-24 -0.2588903 0.038 0.211 9.994575e-20 2
## SRSF10.2 2.845334e-24 0.1409002 0.180 0.467 1.041449e-19 2
## ALG5 2.977811e-24 -0.2772159 0.041 0.217 1.089938e-19 2
## BCAT2.1 3.046538e-24 -0.7085107 0.018 0.173 1.115094e-19 2
## RAB14.1 3.078014e-24 -0.1049974 0.099 0.321 1.126615e-19 2
## NABP2.1 3.139554e-24 -0.6466305 0.024 0.185 1.149140e-19 2
## GLTP.1 3.147741e-24 -1.3933361 0.017 0.170 1.152136e-19 2
## CDKN2AIPNL 3.239479e-24 -3.2542689 0.006 0.149 1.185714e-19 2
## PDHA1 3.255020e-24 -0.6523213 0.044 0.222 1.191402e-19 2
## SYNE2 3.308002e-24 1.5045207 0.595 0.701 1.210795e-19 2
## RRP7A.1 3.363944e-24 -0.9046108 0.035 0.204 1.231271e-19 2
## PABPC4.2 3.379622e-24 0.1233840 0.057 0.248 1.237009e-19 2
## DIMT1 3.538280e-24 -0.4278059 0.048 0.229 1.295081e-19 2
## COPS2.1 3.581251e-24 0.1361864 0.075 0.280 1.310810e-19 2
## COA8 3.666779e-24 -1.4599423 0.018 0.172 1.342115e-19 2
## SELENOS 3.838133e-24 -0.3608131 0.047 0.227 1.404833e-19 2
## ARL6IP6.1 3.856241e-24 -0.1114809 0.080 0.287 1.411461e-19 2
## ISOC2.1 3.897750e-24 -0.9358534 0.020 0.176 1.426654e-19 2
## SNX20.1 4.142127e-24 -0.1627201 0.071 0.270 1.516101e-19 2
## IMMT.1 4.143788e-24 0.1623127 0.126 0.368 1.516709e-19 2
## DCK 4.167850e-24 -0.1966914 0.068 0.265 1.525516e-19 2
## ACTR3.1 4.454741e-24 -0.1323376 0.359 0.762 1.630524e-19 2
## MRPL50 4.466136e-24 -1.5375659 0.012 0.161 1.634695e-19 2
## UQCC3 4.500503e-24 -1.1890560 0.012 0.161 1.647274e-19 2
## RPUSD3 4.626565e-24 -0.4579692 0.036 0.206 1.693415e-19 2
## UBA1 5.118727e-24 -0.1924696 0.062 0.252 1.873556e-19 2
## RNF114 5.174277e-24 -0.1200537 0.053 0.238 1.893889e-19 2
## SYPL1.2 5.210612e-24 -0.6294049 0.032 0.198 1.907188e-19 2
## ACD.1 5.251571e-24 -1.3061721 0.015 0.166 1.922180e-19 2
## UBE2S.1 5.615819e-24 0.1738986 0.155 0.418 2.055502e-19 2
## FLYWCH2 6.040556e-24 -0.8681579 0.024 0.183 2.210964e-19 2
## EVI2A.1 6.081957e-24 -0.3699145 0.060 0.250 2.226118e-19 2
## SLC66A3.1 6.090490e-24 -0.4453246 0.047 0.225 2.229241e-19 2
## MRPL42 6.172182e-24 -0.7769149 0.041 0.214 2.259142e-19 2
## NINJ1.1 7.197350e-24 -1.9522010 0.011 0.157 2.634374e-19 2
## ARL8A 7.251714e-24 -1.0448247 0.035 0.202 2.654272e-19 2
## FKBP4.2 7.307203e-24 -0.4934410 0.030 0.194 2.674583e-19 2
## BRIX1.2 7.326888e-24 -0.8737344 0.026 0.186 2.681787e-19 2
## YWHAE.2 7.621402e-24 0.1672656 0.110 0.342 2.789585e-19 2
## CCR7.2 7.856464e-24 -1.2080120 0.030 0.192 2.875623e-19 2
## CDC34.1 8.017740e-24 -0.4018016 0.045 0.222 2.934653e-19 2
## DENR 8.674767e-24 0.1463169 0.083 0.291 3.175138e-19 2
## CDC42SE1.2 8.741454e-24 0.2269225 0.202 0.504 3.199547e-19 2
## BTG3 9.940690e-24 -0.5110200 0.030 0.193 3.638491e-19 2
## SLU7.1 1.026757e-23 -0.4883733 0.051 0.233 3.758135e-19 2
## GRPEL1.1 1.049164e-23 -0.4077369 0.041 0.213 3.840150e-19 2
## NFU1 1.067442e-23 -1.2202121 0.017 0.168 3.907050e-19 2
## ARMCX6 1.113317e-23 -1.6128570 0.017 0.167 4.074963e-19 2
## UBQLN2.2 1.138746e-23 -0.2200471 0.069 0.263 4.168040e-19 2
## POLR2J.1 1.219350e-23 -1.0940678 0.018 0.170 4.463064e-19 2
## OPTN.2 1.235557e-23 0.3166513 0.217 0.537 4.522384e-19 2
## CBX1.2 1.250143e-23 -0.5720906 0.036 0.204 4.575775e-19 2
## CHCHD7 1.319541e-23 -0.5371019 0.035 0.201 4.829784e-19 2
## MTX1 1.389706e-23 -0.4395543 0.042 0.215 5.086603e-19 2
## CD2BP2.1 1.393134e-23 -0.4732667 0.042 0.215 5.099149e-19 2
## SAP30BP 1.394619e-23 0.1433858 0.075 0.276 5.104583e-19 2
## SGF29 1.431783e-23 -1.0784019 0.021 0.176 5.240612e-19 2
## ARMC10 1.435462e-23 -0.6287133 0.036 0.204 5.254077e-19 2
## CD69 1.452630e-23 1.3470967 0.671 0.797 5.316915e-19 2
## TRMT10C.1 1.455476e-23 -0.8569217 0.032 0.195 5.327332e-19 2
## LLPH 1.479173e-23 -0.9366589 0.027 0.187 5.414070e-19 2
## XRN2.1 1.538679e-23 0.1794191 0.165 0.440 5.631872e-19 2
## TMED5.1 1.548993e-23 0.1054319 0.098 0.316 5.669624e-19 2
## SHFL.1 1.553491e-23 0.3873902 0.102 0.327 5.686088e-19 2
## EPB41L4A-AS1 1.569452e-23 -1.3696996 0.020 0.172 5.744508e-19 2
## EXOSC9.1 1.572903e-23 -0.2963632 0.041 0.213 5.757141e-19 2
## FAM204A 1.577133e-23 -0.2112864 0.063 0.254 5.772621e-19 2
## PSMG3.1 1.625304e-23 -0.3115042 0.027 0.187 5.948936e-19 2
## YARS.2 1.680000e-23 0.2355556 0.116 0.350 6.149138e-19 2
## YDJC 1.731001e-23 -0.4507198 0.041 0.212 6.335811e-19 2
## CD79B 1.753789e-23 -1.2507233 0.017 0.166 6.419219e-19 2
## MRPS31 1.779465e-23 -0.4246945 0.050 0.228 6.513197e-19 2
## COX11 1.819924e-23 -0.9871515 0.021 0.175 6.661285e-19 2
## BLVRA.2 1.952544e-23 -1.7574436 0.009 0.151 7.146702e-19 2
## SRSF6.1 2.045169e-23 -0.2556451 0.059 0.244 7.485729e-19 2
## DLD.1 2.079259e-23 -0.1113046 0.056 0.239 7.610504e-19 2
## YEATS4.2 2.149731e-23 -0.8918113 0.035 0.200 7.868445e-19 2
## GTPBP4 2.204514e-23 -0.3000153 0.051 0.231 8.068961e-19 2
## SMIM7.1 2.225549e-23 -0.1217808 0.078 0.281 8.145955e-19 2
## ZMAT5 2.305146e-23 -1.5265986 0.014 0.160 8.437294e-19 2
## LAMTOR3 2.413403e-23 -0.5808169 0.038 0.205 8.833536e-19 2
## PRMT2.2 2.455798e-23 0.2595525 0.247 0.587 8.988710e-19 2
## TGFB1.2 2.636262e-23 0.1174001 0.313 0.695 9.649248e-19 2
## MRPS17.1 2.785397e-23 -1.0303218 0.014 0.160 1.019511e-18 2
## MRPS18A 2.810209e-23 -0.9643898 0.023 0.177 1.028593e-18 2
## SAT2.1 2.847490e-23 -0.5562376 0.045 0.219 1.042238e-18 2
## IL27RA 2.863031e-23 0.1425079 0.107 0.332 1.047926e-18 2
## DNTTIP2.1 2.946769e-23 0.1982319 0.089 0.298 1.078577e-18 2
## MRPL46.1 2.947666e-23 -1.7308142 0.012 0.156 1.078905e-18 2
## CDCA7.2 3.120633e-23 -0.5416743 0.030 0.191 1.142214e-18 2
## PLPBP 3.147686e-23 -1.1671472 0.018 0.168 1.152116e-18 2
## SLC52A2.1 3.437273e-23 -0.5083301 0.033 0.196 1.258111e-18 2
## ATF6B.1 3.549517e-23 0.2210330 0.107 0.330 1.299194e-18 2
## DRG2 3.568854e-23 -0.4814550 0.036 0.202 1.306272e-18 2
## CKS1B.2 3.655095e-23 -0.6710633 0.029 0.187 1.337838e-18 2
## GEMIN7.1 3.884687e-23 -0.7972196 0.017 0.165 1.421873e-18 2
## AP1S2 4.183323e-23 -0.1211583 0.027 0.185 1.531180e-18 2
## PHAX.1 4.199298e-23 -0.3854836 0.063 0.250 1.537027e-18 2
## SNRNP70 4.422928e-23 0.3891531 0.164 0.435 1.618880e-18 2
## GAMT.2 4.462012e-23 -0.7139643 0.018 0.167 1.633185e-18 2
## DYNLT3.1 4.500845e-23 -1.0034868 0.023 0.176 1.647399e-18 2
## SDHA 4.549223e-23 0.2610874 0.146 0.403 1.665107e-18 2
## COX20 4.601034e-23 -0.6796635 0.032 0.192 1.684071e-18 2
## RAD51C.1 4.661306e-23 -1.0622592 0.020 0.170 1.706131e-18 2
## WARS 4.729636e-23 -0.3243927 0.060 0.245 1.731141e-18 2
## API5 4.780065e-23 0.1155276 0.080 0.281 1.749599e-18 2
## MAGOHB 4.853905e-23 -0.9768535 0.029 0.186 1.776626e-18 2
## SLC39A3 4.860811e-23 -1.9603199 0.011 0.152 1.779154e-18 2
## DDX39A.2 5.018844e-23 0.3426046 0.168 0.438 1.836997e-18 2
## COPS4 5.069265e-23 -0.5968876 0.036 0.200 1.855452e-18 2
## HTRA2 5.136206e-23 -0.7688084 0.030 0.190 1.879954e-18 2
## HRAS.1 5.395172e-23 -0.3090873 0.027 0.184 1.974741e-18 2
## C1orf162.2 5.401830e-23 -2.0358798 0.011 0.152 1.977178e-18 2
## RABL6 5.549911e-23 0.4234240 0.134 0.381 2.031378e-18 2
## C19orf48.2 5.891726e-23 -0.3143937 0.035 0.197 2.156490e-18 2
## TMEM250 6.169275e-23 -0.6281404 0.033 0.195 2.258078e-18 2
## AIMP2 6.311051e-23 -1.1506111 0.018 0.166 2.309971e-18 2
## CRTAP 6.682522e-23 -0.8085333 0.024 0.178 2.445937e-18 2
## NGDN 6.745626e-23 -0.3301218 0.047 0.220 2.469034e-18 2
## EFHD2.2 7.133373e-23 0.1883058 0.156 0.418 2.610957e-18 2
## HNRNPUL2.1 7.516296e-23 0.2042140 0.155 0.416 2.751115e-18 2
## TMEM70.1 7.525430e-23 -1.0645126 0.021 0.171 2.754458e-18 2
## AGPAT2.2 7.678663e-23 -1.9539624 0.011 0.151 2.810544e-18 2
## ENOPH1 7.991651e-23 -0.1808078 0.042 0.211 2.925104e-18 2
## FPGS 8.308441e-23 0.2728876 0.083 0.286 3.041056e-18 2
## HSPA9.1 8.491137e-23 0.1996071 0.198 0.493 3.107926e-18 2
## PCLAF.2 8.937233e-23 -2.3402232 0.015 0.158 3.271206e-18 2
## MCM5.2 8.994421e-23 -0.8185451 0.042 0.208 3.292138e-18 2
## ARRDC1.1 9.519074e-23 -0.2361095 0.048 0.222 3.484172e-18 2
## WDR45.2 9.609819e-23 -0.5492541 0.036 0.199 3.517386e-18 2
## RWDD4 9.953764e-23 -1.8049882 0.014 0.156 3.643277e-18 2
## SURF2 1.037066e-22 -1.0017384 0.018 0.165 3.795868e-18 2
## TOMM40.1 1.096267e-22 -0.3095189 0.051 0.226 4.012555e-18 2
## UBL4A 1.105070e-22 -0.5310141 0.035 0.196 4.044778e-18 2
## RING1.1 1.250358e-22 -0.5651790 0.035 0.196 4.576560e-18 2
## LBR 1.345022e-22 0.2237560 0.189 0.481 4.923050e-18 2
## RHNO1 1.346025e-22 -2.1104464 0.009 0.147 4.926720e-18 2
## HAT1.2 1.368872e-22 -0.5635387 0.039 0.204 5.010345e-18 2
## GRN.1 1.483572e-22 -0.3543811 0.057 0.236 5.430172e-18 2
## COA5 1.502161e-22 -0.7383651 0.027 0.182 5.498211e-18 2
## HAUS4.2 1.531134e-22 -0.8526748 0.023 0.173 5.604256e-18 2
## FAM177A1.1 1.571220e-22 0.2867853 0.089 0.295 5.750981e-18 2
## GSS.1 1.601528e-22 -0.8295407 0.035 0.195 5.861912e-18 2
## RBM38.1 1.619265e-22 -0.3796272 0.042 0.209 5.926833e-18 2
## TCEAL8 1.718308e-22 -1.0125795 0.023 0.173 6.289352e-18 2
## PMVK.1 1.749029e-22 0.3083629 0.077 0.273 6.401797e-18 2
## PXMP2.2 1.771392e-22 -1.4556542 0.012 0.152 6.483650e-18 2
## PTPRC.2 1.788495e-22 0.8462902 0.708 0.897 6.546248e-18 2
## CLPTM1L.1 1.854879e-22 -0.1253716 0.057 0.236 6.789229e-18 2
## UNG.2 1.907834e-22 -1.5672002 0.012 0.152 6.983054e-18 2
## EIF4EBP2.1 1.929297e-22 0.1164491 0.081 0.280 7.061613e-18 2
## AKTIP.1 1.952893e-22 -0.4764184 0.053 0.227 7.147977e-18 2
## DYNLT1.1 2.020581e-22 0.5872136 0.114 0.341 7.395731e-18 2
## MTCH1 2.023884e-22 0.4116921 0.120 0.352 7.407820e-18 2
## MRTO4.2 2.069591e-22 -0.9086814 0.018 0.164 7.575119e-18 2
## STARD7 2.122434e-22 0.1591648 0.080 0.277 7.768534e-18 2
## NUDT2 2.135826e-22 0.3314442 0.054 0.231 7.817550e-18 2
## RGS14.1 2.177884e-22 0.3543127 0.108 0.329 7.971490e-18 2
## CEBPZOS.1 2.222385e-22 -0.5709522 0.032 0.189 8.134373e-18 2
## LYRM2 2.382803e-22 -0.1739046 0.051 0.225 8.721537e-18 2
## C11orf68.1 2.387623e-22 -1.1120668 0.024 0.175 8.739179e-18 2
## FN3KRP.2 2.479032e-22 -1.3848815 0.017 0.160 9.073753e-18 2
## ANTKMT.2 2.525344e-22 -1.2707217 0.017 0.160 9.243264e-18 2
## ATPAF1.1 2.535839e-22 -0.1034042 0.056 0.233 9.281679e-18 2
## CFAP36 2.739491e-22 -0.5106634 0.036 0.197 1.002708e-17 2
## ARL1 2.823526e-22 -0.3328564 0.032 0.189 1.033467e-17 2
## CKAP2.1 2.853577e-22 -0.6245562 0.038 0.199 1.044466e-17 2
## PRKAG1 2.894584e-22 -0.4376918 0.036 0.196 1.059476e-17 2
## CDK5 2.932208e-22 -2.3954101 0.009 0.145 1.073247e-17 2
## TESC.2 3.000957e-22 -0.4688085 0.045 0.213 1.098410e-17 2
## HSF1 3.044622e-22 -0.1650760 0.060 0.240 1.114393e-17 2
## PTBP1 3.124275e-22 0.3871370 0.126 0.362 1.143547e-17 2
## SLIRP 3.137908e-22 -0.1056719 0.054 0.230 1.148537e-17 2
## LIX1L 3.413893e-22 -0.3877645 0.047 0.215 1.249553e-17 2
## NOA1 3.504451e-22 -0.7242064 0.032 0.188 1.282699e-17 2
## TMEM203 3.507234e-22 -0.7423747 0.039 0.201 1.283718e-17 2
## ARF3.2 3.594149e-22 -0.1747862 0.059 0.237 1.315530e-17 2
## RAD23B 3.912225e-22 0.5215522 0.138 0.382 1.431953e-17 2
## APIP 4.055129e-22 -0.6828422 0.027 0.179 1.484258e-17 2
## PLRG1 4.581017e-22 -0.4400464 0.038 0.198 1.676744e-17 2
## OGFR.1 4.644072e-22 0.2427556 0.080 0.275 1.699823e-17 2
## PSME3.1 5.057646e-22 -0.1161464 0.060 0.239 1.851199e-17 2
## RRAGA.1 5.221871e-22 -0.3743129 0.038 0.198 1.911309e-17 2
## CHMP1A 5.228504e-22 -0.3497033 0.051 0.222 1.913737e-17 2
## COMMD1 5.363715e-22 0.1865371 0.090 0.294 1.963227e-17 2
## SINHCAF 5.375529e-22 0.2392875 0.095 0.302 1.967551e-17 2
## NAE1.1 5.555272e-22 -0.1608471 0.050 0.220 2.033341e-17 2
## HDHD5.1 5.646215e-22 -0.7062825 0.029 0.181 2.066628e-17 2
## PDCD10.1 5.724524e-22 0.2293751 0.122 0.351 2.095290e-17 2
## DCUN1D5 6.136016e-22 -0.2246627 0.047 0.214 2.245904e-17 2
## EXOSC4.2 6.693415e-22 -2.3454204 0.006 0.137 2.449924e-17 2
## DANCR.1 7.046345e-22 0.1459993 0.056 0.230 2.579103e-17 2
## MAD1L1.1 7.174876e-22 0.4442665 0.152 0.405 2.626148e-17 2
## SEPTIN6 7.529172e-22 0.1572320 0.296 0.672 2.755828e-17 2
## PSMG4 7.610179e-22 -0.3306802 0.029 0.181 2.785478e-17 2
## PAF1 7.744849e-22 -0.2619889 0.042 0.205 2.834769e-17 2
## PIM3.1 8.077965e-22 -0.1039849 0.050 0.219 2.956697e-17 2
## FAM120AOS 8.563002e-22 -0.3815718 0.041 0.202 3.134230e-17 2
## TYMS.2 8.624991e-22 -1.8053074 0.035 0.186 3.156919e-17 2
## MRPL35.1 8.714941e-22 -1.0496833 0.012 0.148 3.189843e-17 2
## INPP4B.2 8.735268e-22 1.6670492 0.591 0.702 3.197283e-17 2
## THAP7.1 8.736434e-22 -0.5084405 0.024 0.172 3.197710e-17 2
## CDKN1B.2 8.777911e-22 0.1365036 0.146 0.387 3.212891e-17 2
## DPM3.1 8.808351e-22 -0.4576970 0.024 0.171 3.224033e-17 2
## ZDHHC24.1 8.983361e-22 -0.5157263 0.036 0.194 3.288090e-17 2
## C1GALT1C1.1 9.387831e-22 -0.6514111 0.026 0.174 3.436134e-17 2
## CHID1 9.986517e-22 -1.1741389 0.018 0.160 3.655265e-17 2
## SDR39U1 1.054077e-21 -0.3741643 0.047 0.213 3.858133e-17 2
## C15orf40 1.067266e-21 -0.5663627 0.036 0.193 3.906405e-17 2
## PSAP.2 1.070174e-21 0.1893193 0.214 0.514 3.917053e-17 2
## PARL 1.144827e-21 -0.5435767 0.044 0.207 4.190295e-17 2
## MYD88.1 1.162067e-21 -0.3822733 0.039 0.199 4.253397e-17 2
## SPSB3.1 1.307257e-21 0.2874546 0.111 0.327 4.784822e-17 2
## SNHG5 1.323537e-21 0.6543553 0.203 0.500 4.844408e-17 2
## STYXL1 1.355257e-21 -0.2223648 0.035 0.191 4.960513e-17 2
## LAG3.2 1.451889e-21 -0.5111058 0.060 0.234 5.314205e-17 2
## RAB33A.1 1.461418e-21 -0.7038853 0.017 0.156 5.349084e-17 2
## NAPRT 1.530087e-21 -0.4809282 0.039 0.198 5.600423e-17 2
## SRSF8 1.613813e-21 -0.7381629 0.042 0.203 5.906878e-17 2
## DNAJB1.1 1.793484e-21 0.4748177 0.188 0.465 6.564512e-17 2
## PTGES2 1.809649e-21 -0.3708087 0.042 0.203 6.623679e-17 2
## PPP6R1.1 1.816434e-21 0.1918736 0.119 0.342 6.648511e-17 2
## CD40LG.2 1.819729e-21 -0.4189354 0.063 0.238 6.660572e-17 2
## SH3GLB1.1 1.826563e-21 0.2310813 0.095 0.298 6.685585e-17 2
## TMEM60.1 1.847759e-21 -0.6311646 0.029 0.178 6.763169e-17 2
## SAE1.2 2.004781e-21 0.3505457 0.111 0.326 7.337898e-17 2
## TCEA2 2.006313e-21 -0.6196113 0.026 0.172 7.343506e-17 2
## PARVG.1 2.033601e-21 0.3205050 0.131 0.363 7.443385e-17 2
## CDIP1.1 2.035016e-21 -1.2648888 0.023 0.166 7.448565e-17 2
## HEXB.1 2.156553e-21 -0.4745934 0.030 0.180 7.893415e-17 2
## CTDSP1.1 2.170710e-21 0.1653394 0.099 0.306 7.945235e-17 2
## H3F3B.1 2.209594e-21 -0.2834732 0.621 0.909 8.087556e-17 2
## CHTOP 2.233836e-21 -0.4412405 0.056 0.227 8.176286e-17 2
## TMX2.1 2.236155e-21 -0.3229611 0.045 0.208 8.184775e-17 2
## MRPL44.1 2.260518e-21 -1.1921081 0.026 0.172 8.273946e-17 2
## NAGK 2.363106e-21 -0.4913296 0.039 0.197 8.649442e-17 2
## PEPD.1 2.401133e-21 -0.1454991 0.056 0.227 8.788626e-17 2
## PIGP 2.402240e-21 -0.9747897 0.014 0.149 8.792680e-17 2
## PRPF38B 2.515595e-21 0.4562738 0.168 0.432 9.207582e-17 2
## SERINC1.2 2.581239e-21 0.1562155 0.137 0.374 9.447852e-17 2
## AL133415.1 2.598958e-21 -0.9107292 0.017 0.154 9.512707e-17 2
## NIP7 2.665857e-21 -0.7775839 0.026 0.171 9.757571e-17 2
## SYTL1.2 2.701481e-21 0.4334020 0.138 0.376 9.887960e-17 2
## ID2.2 2.704622e-21 0.2432078 0.269 0.600 9.899458e-17 2
## MSN.1 2.728262e-21 0.1122483 0.334 0.725 9.985985e-17 2
## USP14.1 2.784029e-21 0.2175793 0.090 0.289 1.019010e-16 2
## CD164.1 2.800546e-21 0.2611756 0.226 0.540 1.025056e-16 2
## SDHAF3 2.801832e-21 -0.9026693 0.020 0.160 1.025527e-16 2
## GGNBP2 2.833637e-21 0.2415457 0.152 0.398 1.037168e-16 2
## RAB2A 2.854783e-21 0.3347494 0.171 0.436 1.044908e-16 2
## TP53TG1.1 2.917657e-21 -1.7987761 0.012 0.145 1.067921e-16 2
## SLC39A7 2.974255e-21 -0.7259893 0.032 0.182 1.088637e-16 2
## UBE2R2 3.038749e-21 0.2130694 0.101 0.309 1.112243e-16 2
## TOR3A 3.053879e-21 -0.4969724 0.039 0.196 1.117781e-16 2
## IL16.2 3.102327e-21 0.1993796 0.244 0.565 1.135514e-16 2
## HMG20B 3.109285e-21 -0.2270502 0.036 0.191 1.138061e-16 2
## NANS.2 3.133649e-21 0.1473225 0.059 0.232 1.146978e-16 2
## EXOSC3.1 3.400873e-21 -0.6392250 0.033 0.184 1.244788e-16 2
## SERTAD1.1 3.675855e-21 0.5526541 0.096 0.300 1.345436e-16 2
## LSM12 3.739538e-21 -0.1820196 0.047 0.210 1.368746e-16 2
## TMEM106B 3.800911e-21 0.1720579 0.083 0.274 1.391209e-16 2
## NPDC1.1 4.224312e-21 -0.4301109 0.051 0.216 1.546183e-16 2
## YIPF5 4.224590e-21 -0.2916291 0.038 0.193 1.546284e-16 2
## TMEM11.1 4.243059e-21 -0.8886688 0.017 0.153 1.553045e-16 2
## RPA1 4.348943e-21 0.2342476 0.146 0.387 1.591800e-16 2
## MTHFD2.2 4.376042e-21 0.4161134 0.125 0.348 1.601719e-16 2
## MYL6B.1 4.410857e-21 -1.3395170 0.011 0.141 1.614462e-16 2
## SLC39A1.1 4.846219e-21 -1.9313087 0.008 0.135 1.773813e-16 2
## RAB4B.1 4.921801e-21 0.1643994 0.062 0.236 1.801477e-16 2
## GMPPA 4.999553e-21 -0.7568421 0.015 0.150 1.829936e-16 2
## CASP1.1 5.091742e-21 -0.5799622 0.039 0.195 1.863679e-16 2
## MCUR1.1 5.226465e-21 -0.3866352 0.030 0.178 1.912991e-16 2
## GUSB.1 5.232073e-21 0.1654580 0.069 0.250 1.915043e-16 2
## SMIM15.1 5.476458e-21 -0.1554266 0.048 0.211 2.004493e-16 2
## GMNN.2 5.616046e-21 -1.2265294 0.020 0.158 2.055585e-16 2
## MVB12A 5.887086e-21 -2.1223508 0.012 0.144 2.154791e-16 2
## AGGF1 5.902872e-21 0.2958478 0.081 0.271 2.160569e-16 2
## IFI30.1 6.061613e-21 -0.8935371 0.021 0.161 2.218671e-16 2
## FOPNL.1 6.124498e-21 -0.9168666 0.020 0.158 2.241689e-16 2
## SNW1 6.869043e-21 0.3746767 0.090 0.287 2.514207e-16 2
## PTMS.1 6.982084e-21 -0.5433777 0.050 0.212 2.555582e-16 2
## BORCS7 7.143271e-21 -0.5275954 0.029 0.175 2.614580e-16 2
## TOMM70 7.211936e-21 -0.4722811 0.042 0.199 2.639713e-16 2
## HIRIP3.2 7.276687e-21 -1.1874122 0.012 0.143 2.663413e-16 2
## TRABD.1 7.491161e-21 0.5809256 0.182 0.455 2.741915e-16 2
## SCO2 7.633452e-21 -0.3367105 0.030 0.178 2.793996e-16 2
## FAM104A.1 7.770002e-21 -0.1970089 0.048 0.211 2.843976e-16 2
## HACD3.1 8.212074e-21 -0.2561530 0.048 0.210 3.005783e-16 2
## BLVRB.1 8.578635e-21 -0.6707675 0.021 0.160 3.139952e-16 2
## LMNB1.2 8.742374e-21 -0.1023206 0.071 0.250 3.199884e-16 2
## CDC14A.1 8.808505e-21 1.9294143 0.501 0.524 3.224089e-16 2
## ENDOG 9.145505e-21 -1.1144301 0.014 0.146 3.347438e-16 2
## LARS 9.228901e-21 0.2715948 0.116 0.331 3.377963e-16 2
## BTN3A2.2 9.733526e-21 0.2758565 0.105 0.313 3.562665e-16 2
## RFC2.2 1.008947e-20 -0.8778683 0.018 0.154 3.692949e-16 2
## MT1X.1 1.019773e-20 -0.8277820 0.017 0.151 3.732573e-16 2
## ATP6V1D.1 1.023466e-20 -0.1881638 0.050 0.213 3.746090e-16 2
## SMARCC2 1.058114e-20 0.2880317 0.122 0.342 3.872908e-16 2
## ARFGAP3 1.177086e-20 0.1477932 0.086 0.277 4.308372e-16 2
## PEX16.1 1.211282e-20 0.1859940 0.072 0.252 4.433533e-16 2
## IMPDH1 1.234094e-20 -0.7205765 0.033 0.182 4.517032e-16 2
## FAM200B 1.254978e-20 -1.7742172 0.012 0.142 4.593471e-16 2
## TNRC6B.1 1.302388e-20 1.9375475 0.469 0.460 4.766999e-16 2
## VTA1 1.305298e-20 -0.1034948 0.060 0.232 4.777652e-16 2
## UTP11.1 1.329350e-20 -1.0751313 0.017 0.150 4.865688e-16 2
## GLS 1.345016e-20 1.8471374 0.516 0.562 4.923027e-16 2
## TACC3.1 1.349971e-20 0.1844995 0.086 0.276 4.941164e-16 2
## FTSJ3.1 1.383946e-20 -0.3135106 0.036 0.187 5.065521e-16 2
## DNAJC21.1 1.417792e-20 0.2734361 0.099 0.301 5.189402e-16 2
## SNX15 1.476570e-20 -1.0343458 0.021 0.159 5.404543e-16 2
## SMC2.2 1.541311e-20 -0.3049122 0.042 0.196 5.641507e-16 2
## ACADM 1.573230e-20 -0.3195186 0.056 0.221 5.758338e-16 2
## GLIPR1.1 1.580043e-20 0.1097860 0.125 0.343 5.783272e-16 2
## HSPA1A.1 1.583300e-20 -0.2292878 0.069 0.245 5.795195e-16 2
## KRCC1 1.663671e-20 -0.4890127 0.057 0.223 6.089367e-16 2
## IRF3 1.759572e-20 0.3875393 0.087 0.278 6.440387e-16 2
## ACOT13 1.826840e-20 -1.1301923 0.020 0.156 6.686598e-16 2
## MRPS5 1.951612e-20 0.2838182 0.092 0.287 7.143289e-16 2
## NKAP 1.958496e-20 -0.1591862 0.041 0.195 7.168486e-16 2
## SEC61A1 1.962816e-20 0.3031835 0.129 0.355 7.184300e-16 2
## SMIM3.2 1.964171e-20 -0.6081193 0.033 0.180 7.189259e-16 2
## SLC25A20.1 2.021345e-20 -0.7523442 0.024 0.164 7.398527e-16 2
## ERCC1 2.071410e-20 0.6735183 0.110 0.320 7.581773e-16 2
## CAPRIN1.1 2.105557e-20 0.2540049 0.119 0.334 7.706761e-16 2
## AOAH.2 2.140381e-20 2.3864888 0.368 0.281 7.834224e-16 2
## GPN1 2.194490e-20 -0.8750815 0.029 0.171 8.032273e-16 2
## BNIP2 2.231217e-20 0.1504129 0.113 0.323 8.166700e-16 2
## PIN4.2 2.297193e-20 0.1767238 0.044 0.200 8.408187e-16 2
## DEDD2.1 2.521258e-20 -0.2144012 0.060 0.228 9.228309e-16 2
## ELP6.1 2.651542e-20 -0.4939557 0.029 0.172 9.705174e-16 2
## NME4.2 2.676712e-20 -2.4754585 0.005 0.125 9.797301e-16 2
## PTPMT1.1 2.901320e-20 -1.1110564 0.018 0.151 1.061941e-15 2
## CD151.1 3.087852e-20 -0.3896078 0.035 0.182 1.130216e-15 2
## CYB5A.2 3.289642e-20 -2.1325141 0.008 0.130 1.204075e-15 2
## GPKOW.1 3.444876e-20 -1.0844305 0.018 0.151 1.260894e-15 2
## VRK1.2 3.474326e-20 -0.3472907 0.047 0.203 1.271673e-15 2
## TSFM.1 3.512960e-20 -0.9382691 0.023 0.160 1.285814e-15 2
## TMEM222 3.702071e-20 -0.6069502 0.030 0.173 1.355032e-15 2
## COMMD2.1 3.750138e-20 -0.6000223 0.038 0.187 1.372626e-15 2
## DHFR.2 3.838994e-20 -0.6838319 0.027 0.168 1.405148e-15 2
## TTC1 4.066176e-20 0.2771676 0.077 0.258 1.488302e-15 2
## ZPR1 4.291795e-20 -0.3769919 0.036 0.184 1.570883e-15 2
## PHF23.1 4.402616e-20 -0.2829559 0.044 0.198 1.611446e-15 2
## GINS2.2 4.554743e-20 -1.5904204 0.014 0.141 1.667127e-15 2
## CENPK.2 4.580020e-20 0.1181069 0.114 0.322 1.676379e-15 2
## MRPL45.1 4.582196e-20 -0.6429664 0.027 0.168 1.677175e-15 2
## GOLPH3.1 4.890477e-20 0.1831509 0.104 0.305 1.790012e-15 2
## INPP5K.1 4.892501e-20 -0.3627781 0.042 0.195 1.790753e-15 2
## TCTEX1D2.1 4.932833e-20 -0.2567529 0.032 0.176 1.805516e-15 2
## TOLLIP 4.976965e-20 -1.0204298 0.020 0.153 1.821669e-15 2
## ACTL6A.1 5.084920e-20 -0.4030825 0.036 0.184 1.861182e-15 2
## SMPD1.1 5.143106e-20 -0.1832108 0.035 0.181 1.882480e-15 2
## LZIC 5.218106e-20 -0.1578296 0.038 0.187 1.909931e-15 2
## NPRL2.1 5.287468e-20 -0.1295574 0.051 0.211 1.935319e-15 2
## TPP1.2 5.293992e-20 0.2143152 0.104 0.304 1.937707e-15 2
## SMAD3.1 5.352071e-20 2.3717703 0.409 0.353 1.958965e-15 2
## MED4 5.497830e-20 0.2467605 0.101 0.300 2.012316e-15 2
## C5orf51 6.097813e-20 -0.3788763 0.036 0.183 2.231922e-15 2
## AAAS.1 6.233871e-20 -0.7701742 0.027 0.167 2.281721e-15 2
## AATF 6.273510e-20 0.5192751 0.120 0.336 2.296230e-15 2
## TBPL1 6.337323e-20 -0.6167301 0.029 0.169 2.319587e-15 2
## NSMCE4A.1 6.909286e-20 -0.6795639 0.036 0.182 2.528937e-15 2
## CINP 7.039261e-20 -0.7278607 0.018 0.149 2.576510e-15 2
## GFER 7.187499e-20 -0.4002740 0.027 0.166 2.630768e-15 2
## HSPBP1 7.232176e-20 -0.6590515 0.018 0.149 2.647121e-15 2
## EMC8 7.268167e-20 -0.5429705 0.024 0.161 2.660294e-15 2
## VAMP7.1 7.399717e-20 -0.8606885 0.026 0.163 2.708445e-15 2
## MAP4K1.1 7.964727e-20 0.5319486 0.146 0.381 2.915249e-15 2
## CDCA4.2 8.302818e-20 -1.0452698 0.021 0.154 3.038998e-15 2
## SRRT.1 9.178091e-20 0.3629384 0.095 0.287 3.359365e-15 2
## MRPL39 9.432779e-20 -0.6988625 0.027 0.166 3.452586e-15 2
## QTRT1.1 9.529419e-20 -0.2000953 0.041 0.191 3.487958e-15 2
## NUP37.2 9.801584e-20 -0.5432114 0.032 0.174 3.587576e-15 2
## SVBP 1.042601e-19 -0.8526773 0.015 0.143 3.816127e-15 2
## RABGGTB 1.062270e-19 -0.2810651 0.050 0.206 3.888119e-15 2
## RPP30.1 1.106516e-19 -0.4789124 0.036 0.182 4.050069e-15 2
## OVCA2.1 1.113153e-19 -2.0743883 0.009 0.130 4.074362e-15 2
## ARFRP1 1.358782e-19 -0.2209848 0.047 0.200 4.973415e-15 2
## IKBIP.1 1.386751e-19 -0.3136600 0.033 0.176 5.075785e-15 2
## TRIM69.2 1.451134e-19 0.3499617 0.089 0.276 5.311439e-15 2
## WDR46 1.481591e-19 -0.2749364 0.033 0.176 5.422920e-15 2
## AK3 1.495912e-19 -0.1996743 0.051 0.208 5.475338e-15 2
## PGM1 1.515789e-19 -0.1681155 0.035 0.178 5.548093e-15 2
## GLRX2 1.534575e-19 -0.9014665 0.020 0.150 5.616851e-15 2
## PAK2.1 1.545442e-19 0.3997333 0.229 0.531 5.656625e-15 2
## PIGS.1 1.610261e-19 -0.5827759 0.036 0.181 5.893878e-15 2
## MTA2 1.621369e-19 0.1144297 0.087 0.271 5.934533e-15 2
## MED16 1.667782e-19 -0.6295778 0.026 0.161 6.104416e-15 2
## C19orf25.1 1.728155e-19 0.1402599 0.059 0.222 6.325394e-15 2
## POLR2D.1 1.764389e-19 -0.8524140 0.024 0.158 6.458018e-15 2
## FBXO6.1 1.821671e-19 -0.9948805 0.018 0.147 6.667682e-15 2
## COASY 1.830855e-19 -0.2975953 0.026 0.161 6.701297e-15 2
## IRF9 1.875857e-19 0.2338505 0.116 0.323 6.866011e-15 2
## ZNF32 2.065317e-19 -2.4028106 0.006 0.123 7.559474e-15 2
## NSUN5 2.071539e-19 0.1056258 0.038 0.183 7.582248e-15 2
## TPD52L2 2.109534e-19 -0.3606422 0.033 0.174 7.721315e-15 2
## IPO5.2 2.268544e-19 -0.2226299 0.060 0.223 8.303323e-15 2
## NBL1 2.362021e-19 -0.3123027 0.054 0.212 8.645469e-15 2
## ACOT8 2.389691e-19 -0.8008614 0.018 0.146 8.746745e-15 2
## THUMPD3 2.430397e-19 -0.5991627 0.035 0.177 8.895738e-15 2
## AGPAT1 2.470855e-19 -1.1692571 0.020 0.149 9.043823e-15 2
## AP002387.2.1 2.484575e-19 -1.1899574 0.017 0.143 9.094040e-15 2
## TES.2 2.688655e-19 0.2936605 0.126 0.340 9.841017e-15 2
## UBAC1 2.848712e-19 -0.5844611 0.021 0.151 1.042686e-14 2
## BCL2L12.2 3.040169e-19 -1.2066060 0.015 0.140 1.112763e-14 2
## CDT1.2 3.075233e-19 -0.9176358 0.020 0.148 1.125597e-14 2
## TARS.1 3.138983e-19 0.1955450 0.062 0.225 1.148930e-14 2
## CCDC22 3.350842e-19 -0.4669063 0.029 0.165 1.226475e-14 2
## ERAL1 3.374262e-19 -0.3287683 0.027 0.162 1.235047e-14 2
## SOCS3.1 3.579240e-19 0.4477432 0.129 0.344 1.310074e-14 2
## POLR2B 3.848648e-19 0.3844790 0.107 0.304 1.408682e-14 2
## RASSF1.2 3.903868e-19 0.3956802 0.153 0.387 1.428894e-14 2
## RBM22.1 3.935733e-19 0.2750301 0.080 0.256 1.440557e-14 2
## IFRD2.1 3.974875e-19 -1.0499277 0.018 0.145 1.454884e-14 2
## TMEM101 4.075973e-19 -0.5585966 0.030 0.167 1.491888e-14 2
## KIF5B.2 4.122170e-19 0.4847312 0.150 0.384 1.508797e-14 2
## CUTC 4.122493e-19 -0.1921928 0.033 0.173 1.508915e-14 2
## EFCAB14.1 4.218627e-19 0.1048669 0.108 0.306 1.544102e-14 2
## GRB2 4.335634e-19 0.4425554 0.218 0.511 1.586929e-14 2
## PEX11B 4.471124e-19 -0.8796440 0.029 0.164 1.636521e-14 2
## TMPO.2 4.538917e-19 0.2052940 0.134 0.348 1.661334e-14 2
## BORCS6 4.594230e-19 -1.5001170 0.014 0.136 1.681580e-14 2
## SCOC 4.795666e-19 -1.6119780 0.009 0.127 1.755310e-14 2
## NELFCD.1 4.853157e-19 -0.3753005 0.033 0.172 1.776352e-14 2
## CAMTA1 4.999011e-19 0.2431914 0.078 0.254 1.829738e-14 2
## C22orf39 5.041026e-19 -0.3467315 0.032 0.170 1.845116e-14 2
## NR2C2AP.2 5.130274e-19 -1.8953972 0.009 0.127 1.877783e-14 2
## ACTR1A 5.468922e-19 0.1908170 0.069 0.236 2.001735e-14 2
## ANKRD39.1 5.670177e-19 -0.1177620 0.048 0.199 2.075398e-14 2
## EMG1 5.842106e-19 -0.3080621 0.036 0.178 2.138328e-14 2
## FASTK 6.015059e-19 0.1157456 0.048 0.199 2.201632e-14 2
## GIPC1 6.027737e-19 0.2722022 0.071 0.240 2.206272e-14 2
## NCF1 6.237571e-19 -0.7535173 0.024 0.155 2.283076e-14 2
## ATP6V1B2.2 7.273897e-19 0.1546970 0.039 0.183 2.662392e-14 2
## TSEN54.1 7.382784e-19 0.4214945 0.089 0.271 2.702246e-14 2
## GPN3.2 7.451744e-19 -0.5670424 0.018 0.143 2.727487e-14 2
## EIF3B.1 7.646804e-19 0.2952755 0.119 0.325 2.798883e-14 2
## MTDH 7.964617e-19 0.2492397 0.256 0.572 2.915209e-14 2
## RABGAP1L 7.973202e-19 1.7909829 0.519 0.579 2.918351e-14 2
## NRM.2 8.311630e-19 -2.5422688 0.003 0.113 3.042223e-14 2
## TAF6.1 8.451729e-19 -0.5007163 0.023 0.152 3.093502e-14 2
## ARMC6 8.507101e-19 -0.8226698 0.017 0.140 3.113769e-14 2
## LINC00667 8.627658e-19 -0.8000296 0.023 0.152 3.157895e-14 2
## RBM4 8.998465e-19 0.1776848 0.087 0.268 3.293618e-14 2
## CCDC57 9.181674e-19 0.5567334 0.095 0.282 3.360676e-14 2
## DNMT1.2 9.238266e-19 0.3617499 0.156 0.388 3.381390e-14 2
## LINC00623.2 9.448567e-19 0.1715003 0.092 0.274 3.458364e-14 2
## G3BP1 9.611624e-19 0.2783799 0.170 0.415 3.518047e-14 2
## EMB.2 9.640544e-19 0.2804707 0.247 0.554 3.528632e-14 2
## PPA2 9.679402e-19 0.2618026 0.095 0.281 3.542855e-14 2
## ACYP1.1 9.991348e-19 -0.5484542 0.026 0.157 3.657033e-14 2
## YBX3.2 1.087841e-18 0.2616519 0.104 0.297 3.981715e-14 2
## MFSD1.1 1.160902e-18 -0.2423009 0.047 0.195 4.249132e-14 2
## IKBKG 1.220633e-18 0.1525030 0.051 0.203 4.467759e-14 2
## LTA 1.315039e-18 -0.5544355 0.042 0.185 4.813306e-14 2
## TXNDC5.1 1.317225e-18 -0.9898881 0.027 0.158 4.821305e-14 2
## BRCC3 1.339787e-18 -0.3562874 0.035 0.173 4.903888e-14 2
## MPHOSPH10 1.345367e-18 0.2437569 0.054 0.208 4.924314e-14 2
## EBAG9 1.363295e-18 -1.1437502 0.015 0.136 4.989932e-14 2
## VPS26B 1.374978e-18 0.2840317 0.078 0.251 5.032693e-14 2
## FIP1L1 1.408994e-18 0.2986111 0.126 0.337 5.157201e-14 2
## CSNK1A1 1.454601e-18 0.5539132 0.206 0.484 5.324131e-14 2
## FAH.1 1.458085e-18 -0.7247785 0.020 0.145 5.336882e-14 2
## FMC1 1.480056e-18 -1.0615202 0.020 0.144 5.417303e-14 2
## UPF3A.2 1.512518e-18 0.4455968 0.132 0.346 5.536119e-14 2
## YY1.1 1.573146e-18 0.4041223 0.161 0.399 5.758028e-14 2
## PEX2 1.573180e-18 -0.1221904 0.045 0.192 5.758152e-14 2
## PPIL3 1.575437e-18 -0.1147750 0.039 0.180 5.766413e-14 2
## DEXI 1.587360e-18 -0.6608861 0.032 0.167 5.810057e-14 2
## NRBF2 1.693575e-18 -0.1947902 0.039 0.180 6.198822e-14 2
## SNHG1.1 1.700608e-18 0.3158430 0.111 0.308 6.224564e-14 2
## CHRAC1 1.727305e-18 -0.1680374 0.047 0.193 6.322283e-14 2
## DPH3 1.747143e-18 0.1195085 0.042 0.185 6.394891e-14 2
## PLPP5 1.748850e-18 -0.6003072 0.033 0.169 6.401142e-14 2
## GSKIP 1.850687e-18 -1.2848858 0.014 0.132 6.773883e-14 2
## SMIM20 1.868460e-18 -0.9397279 0.018 0.141 6.838937e-14 2
## KLF6 1.871237e-18 1.0969174 0.659 0.808 6.849100e-14 2
## GZMK.2 1.884124e-18 -1.4950216 0.053 0.195 6.896269e-14 2
## TMEM170A 1.892666e-18 -0.1721471 0.054 0.207 6.927538e-14 2
## HOXB2.1 1.931704e-18 -0.4766318 0.023 0.150 7.070423e-14 2
## SMC1A.1 1.952880e-18 0.2512649 0.116 0.313 7.147931e-14 2
## TAX1BP3.1 1.960813e-18 -0.9765365 0.009 0.123 7.176967e-14 2
## RASSF7 2.067794e-18 0.2887314 0.062 0.221 7.568540e-14 2
## NOC2L 2.070746e-18 0.1329823 0.059 0.215 7.579344e-14 2
## LRR1.2 2.155490e-18 -0.2653544 0.020 0.144 7.889526e-14 2
## TM2D1.1 2.190047e-18 -0.1107433 0.069 0.233 8.016010e-14 2
## LRBA.1 2.192181e-18 1.9041922 0.478 0.499 8.023821e-14 2
## NEPRO 2.287915e-18 -0.6405164 0.038 0.176 8.374228e-14 2
## CLK3 2.288259e-18 -0.1452866 0.047 0.193 8.375485e-14 2
## SYS1 2.306660e-18 -0.4814945 0.035 0.171 8.442839e-14 2
## CCNC 2.307641e-18 -0.2989217 0.048 0.195 8.446426e-14 2
## ANXA1.2 2.323194e-18 0.1991453 0.370 0.731 8.503356e-14 2
## ZNF639 2.345233e-18 -0.2985763 0.038 0.176 8.584022e-14 2
## MAD2L1BP 2.380362e-18 -0.7134574 0.021 0.146 8.712601e-14 2
## CDC40 2.404573e-18 0.2968726 0.092 0.272 8.801217e-14 2
## METAP2 2.442338e-18 0.3707522 0.071 0.236 8.939447e-14 2
## NIPSNAP2.2 2.479078e-18 -0.2047969 0.041 0.182 9.073922e-14 2
## SMYD2 2.487613e-18 -0.4377174 0.036 0.174 9.105160e-14 2
## MOCS2 2.495672e-18 -1.1791164 0.015 0.135 9.134658e-14 2
## MCM4.2 2.655401e-18 -0.9933601 0.029 0.159 9.719297e-14 2
## SFXN1.1 2.691446e-18 0.3745481 0.171 0.417 9.851230e-14 2
## SGTA.1 2.700836e-18 -0.4233240 0.029 0.160 9.885601e-14 2
## APBA3.1 2.785937e-18 -0.1775654 0.030 0.163 1.019709e-13 2
## UBA3.1 2.794112e-18 0.3765289 0.126 0.334 1.022701e-13 2
## SMARCA5 2.835980e-18 0.4940550 0.140 0.360 1.038025e-13 2
## TOR2A.1 2.907761e-18 -0.7523605 0.027 0.157 1.064299e-13 2
## ZWINT.2 2.948894e-18 -1.5237486 0.011 0.125 1.079354e-13 2
## RGCC.1 2.981339e-18 0.6735232 0.149 0.375 1.091230e-13 2
## METRN.1 3.199020e-18 -1.6111006 0.011 0.125 1.170905e-13 2
## NUB1 3.218944e-18 0.1947542 0.104 0.293 1.178198e-13 2
## GAR1 3.286288e-18 -0.1213395 0.038 0.176 1.202847e-13 2
## LINC02273.1 3.406245e-18 -0.7180820 0.017 0.136 1.246754e-13 2
## DENND6B.1 3.483534e-18 -0.9556951 0.021 0.145 1.275043e-13 2
## TAX1BP1.1 3.575250e-18 0.4395227 0.227 0.520 1.308613e-13 2
## LMF2.1 3.617930e-18 0.3136204 0.083 0.255 1.324235e-13 2
## TRAP1.2 3.666573e-18 0.2322773 0.051 0.200 1.342039e-13 2
## EID2 4.248760e-18 -0.5072628 0.026 0.153 1.555131e-13 2
## GDI1 4.301211e-18 0.4471245 0.080 0.250 1.574329e-13 2
## STK25 4.302374e-18 0.3617382 0.056 0.207 1.574755e-13 2
## TRA2A 4.383436e-18 0.5520135 0.162 0.398 1.604425e-13 2
## DR1 4.400508e-18 0.3037786 0.120 0.321 1.610674e-13 2
## WTAP 4.426405e-18 0.5471008 0.185 0.438 1.620153e-13 2
## CLEC2B.2 4.564736e-18 0.4749918 0.223 0.504 1.670785e-13 2
## ARL4A 4.616310e-18 -0.6862221 0.029 0.158 1.689662e-13 2
## CHMP2B 4.636947e-18 -0.1753454 0.042 0.183 1.697215e-13 2
## RBMX2 4.644138e-18 -0.7388868 0.017 0.136 1.699847e-13 2
## ZSWIM7 4.673616e-18 -0.4549222 0.027 0.155 1.710637e-13 2
## KAT8 4.716483e-18 -0.2392686 0.036 0.172 1.726327e-13 2
## CYB561A3 4.803033e-18 -1.1759081 0.024 0.149 1.758006e-13 2
## ERAP1 5.070151e-18 0.2215286 0.120 0.320 1.855777e-13 2
## GNAI3 5.100606e-18 0.3436893 0.137 0.350 1.866924e-13 2
## SLC35B1.1 5.139927e-18 -0.4929885 0.038 0.174 1.881316e-13 2
## ACAT1 5.385577e-18 0.2044178 0.069 0.231 1.971229e-13 2
## CCNB1IP1 5.549966e-18 -0.8630265 0.021 0.144 2.031399e-13 2
## EMC9.1 5.742981e-18 -1.3149810 0.008 0.118 2.102046e-13 2
## TSNAX 5.752309e-18 -0.1948184 0.054 0.203 2.105460e-13 2
## RASSF5.1 5.888257e-18 0.3852402 0.173 0.418 2.155220e-13 2
## ERVK3-1 5.988564e-18 0.3451488 0.098 0.281 2.191934e-13 2
## CLPTM1.1 6.329333e-18 0.1064004 0.054 0.204 2.316662e-13 2
## DYRK4 6.622635e-18 -0.7172577 0.021 0.144 2.424017e-13 2
## BRD7 6.813044e-18 0.3184430 0.104 0.291 2.493710e-13 2
## SLC35A4 6.988381e-18 -0.5839216 0.029 0.157 2.557887e-13 2
## NIT1 7.155986e-18 -0.9828851 0.017 0.135 2.619234e-13 2
## TRIM8.1 7.282842e-18 0.2623364 0.054 0.204 2.665666e-13 2
## RP9 7.364086e-18 -1.0472651 0.020 0.140 2.695403e-13 2
## EXOSC6 7.407078e-18 -0.1764715 0.042 0.182 2.711139e-13 2
## BTBD6.1 7.829600e-18 -0.4932824 0.021 0.143 2.865790e-13 2
## SNAPC2 7.893483e-18 -1.2785289 0.020 0.140 2.889173e-13 2
## UBE2J1 7.912559e-18 0.3090920 0.078 0.245 2.896155e-13 2
## EIF4E 7.934888e-18 0.3969090 0.084 0.257 2.904328e-13 2
## ANKRD13A 8.021585e-18 -0.2167773 0.056 0.205 2.936060e-13 2
## PPIF.2 8.372872e-18 -0.2835806 0.041 0.179 3.064639e-13 2
## DOK1 8.639298e-18 -1.2141024 0.018 0.137 3.162156e-13 2
## SDHAF1 8.769035e-18 -0.9953625 0.026 0.151 3.209642e-13 2
## KIF2A.1 9.240344e-18 0.3007112 0.260 0.576 3.382151e-13 2
## MRM2 9.336776e-18 -0.2859511 0.021 0.143 3.417447e-13 2
## TYW3 9.618313e-18 -0.1978426 0.051 0.197 3.520495e-13 2
## OSGEP.1 9.919143e-18 0.4648379 0.072 0.235 3.630605e-13 2
## CASP6 1.007183e-17 -1.1274319 0.020 0.139 3.686491e-13 2
## MRPS22 1.035508e-17 -0.1971299 0.033 0.165 3.790166e-13 2
## SNHG15 1.041420e-17 0.5917548 0.101 0.286 3.811805e-13 2
## FABP5.1 1.049054e-17 -0.9005437 0.012 0.125 3.839748e-13 2
## NIPSNAP3A 1.077054e-17 -1.4363948 0.011 0.122 3.942232e-13 2
## ZNF330 1.088455e-17 -0.3724345 0.032 0.162 3.983964e-13 2
## IRF7.1 1.098660e-17 -0.5912074 0.024 0.148 4.021316e-13 2
## CDC5L 1.100212e-17 0.3974757 0.093 0.271 4.026997e-13 2
## LDLRAP1.2 1.101762e-17 -0.6086333 0.026 0.150 4.032668e-13 2
## EPS8L2.1 1.106471e-17 -2.1144105 0.009 0.119 4.049905e-13 2
## MRPS30 1.127147e-17 -0.2426261 0.045 0.186 4.125584e-13 2
## ILF3 1.127888e-17 0.6758600 0.191 0.448 4.128296e-13 2
## TMEM50B 1.178865e-17 0.1560968 0.065 0.221 4.314881e-13 2
## ARMC1 1.225358e-17 -0.3811060 0.029 0.156 4.485057e-13 2
## WDR34.2 1.339603e-17 -2.0448764 0.009 0.118 4.903214e-13 2
## CHMP6.1 1.357166e-17 -0.3606454 0.030 0.159 4.967500e-13 2
## COPB1 1.521074e-17 0.3267420 0.147 0.365 5.567434e-13 2
## NTPCR 1.593202e-17 -0.6941436 0.017 0.133 5.831437e-13 2
## CXXC1.1 1.595347e-17 0.1734557 0.047 0.187 5.839290e-13 2
## PGRMC2 1.603827e-17 -0.3543553 0.036 0.169 5.870327e-13 2
## AGA 1.607268e-17 -0.6445966 0.027 0.152 5.882921e-13 2
## SNAP23 1.621451e-17 0.1048295 0.062 0.214 5.934836e-13 2
## RFC4.2 1.673732e-17 -0.9242370 0.020 0.138 6.126193e-13 2
## TMIGD2.1 1.683319e-17 -1.6372154 0.011 0.121 6.161284e-13 2
## GNPAT 1.687430e-17 -0.3620380 0.038 0.171 6.176332e-13 2
## ATP1B3.2 1.746832e-17 0.5429937 0.129 0.335 6.393756e-13 2
## ARPP19.1 1.758142e-17 0.2534037 0.078 0.244 6.435153e-13 2
## SLC2A3 1.811237e-17 1.4918109 0.576 0.686 6.629491e-13 2
## CCDC86.2 1.843175e-17 -1.3969426 0.012 0.124 6.746390e-13 2
## MIB2.1 1.860594e-17 -0.1683486 0.036 0.168 6.810146e-13 2
## TESPA1.1 1.889046e-17 0.4124143 0.202 0.461 6.914288e-13 2
## CD70.2 1.929954e-17 -0.1672785 0.062 0.212 7.064017e-13 2
## NIPA2 1.945854e-17 -0.3756064 0.044 0.182 7.122213e-13 2
## ZNF622 1.978897e-17 -0.2080349 0.045 0.184 7.243159e-13 2
## DCAF11.1 1.983594e-17 0.3749220 0.083 0.252 7.260350e-13 2
## UNC50 1.988371e-17 -0.2251158 0.035 0.166 7.277835e-13 2
## ARIH2 2.028841e-17 0.2538808 0.095 0.273 7.425963e-13 2
## NOP2 2.074284e-17 -0.3312179 0.038 0.170 7.592295e-13 2
## PHKG2.1 2.123415e-17 -0.1519455 0.042 0.179 7.772122e-13 2
## RAP2B 2.207699e-17 0.1628037 0.074 0.235 8.080621e-13 2
## AC008555.4.1 2.237080e-17 -1.4469486 0.009 0.117 8.188161e-13 2
## BAG6 2.241180e-17 0.2447426 0.083 0.249 8.203169e-13 2
## GPATCH4 2.451076e-17 -0.1505581 0.030 0.157 8.971427e-13 2
## RNASEH2A.2 2.459814e-17 -1.5323546 0.008 0.114 9.003413e-13 2
## RRM2.2 2.476233e-17 -2.0844120 0.024 0.143 9.063509e-13 2
## SP110.1 2.557288e-17 0.5671445 0.208 0.474 9.360186e-13 2
## RAB27A.1 2.566918e-17 0.3705532 0.113 0.302 9.395432e-13 2
## NUP107.2 2.585403e-17 0.1760930 0.062 0.213 9.463094e-13 2
## PSMD10 2.626817e-17 -0.9736099 0.018 0.134 9.614674e-13 2
## PCGF1 2.632596e-17 -1.3405545 0.012 0.123 9.635828e-13 2
## DCAF7 2.641006e-17 0.3670240 0.119 0.313 9.666609e-13 2
## PDXP.1 2.663857e-17 -1.8348741 0.008 0.114 9.750248e-13 2
## GSPT1 2.703232e-17 0.6622367 0.168 0.404 9.894368e-13 2
## LAP3.1 2.754034e-17 0.5051882 0.093 0.268 1.008032e-12 2
## SERPINB6.1 2.784331e-17 -0.6301610 0.027 0.151 1.019121e-12 2
## CLIC3.2 2.880520e-17 -1.7114619 0.020 0.136 1.054328e-12 2
## EEF1AKMT2 2.898171e-17 -0.3060514 0.038 0.170 1.060789e-12 2
## DEDD 3.188792e-17 -0.6010533 0.024 0.145 1.167162e-12 2
## CITED2.2 3.208554e-17 0.4525780 0.176 0.413 1.174395e-12 2
## GON7 3.263647e-17 -0.5678856 0.027 0.151 1.194560e-12 2
## NKAPD1 3.321384e-17 -0.5718827 0.023 0.142 1.215693e-12 2
## BRCA1.2 3.420913e-17 -0.8158233 0.017 0.131 1.252123e-12 2
## NUDT14.2 3.775348e-17 -0.8521516 0.015 0.128 1.381853e-12 2
## ENO2.1 3.804660e-17 0.2121491 0.042 0.178 1.392582e-12 2
## JMJD8 3.830737e-17 -0.7753903 0.026 0.148 1.402126e-12 2
## SLC1A5.1 3.844633e-17 0.3091838 0.062 0.212 1.407212e-12 2
## PIGX.1 3.884548e-17 0.1360853 0.044 0.180 1.421822e-12 2
## MAP7D1.1 3.933195e-17 0.4159818 0.159 0.384 1.439628e-12 2
## CAPN2.2 4.018404e-17 0.4643376 0.217 0.488 1.470816e-12 2
## CBR3.2 4.038342e-17 0.1901040 0.023 0.142 1.478114e-12 2
## CDK1.2 4.059981e-17 -4.0279172 0.002 0.101 1.486034e-12 2
## C1orf131 4.061168e-17 -0.2103075 0.032 0.158 1.486469e-12 2
## DNLZ 4.101276e-17 -1.1590633 0.012 0.122 1.501149e-12 2
## UNC45A.1 4.169074e-17 0.1356915 0.056 0.201 1.525964e-12 2
## BZW2 4.254579e-17 0.3211295 0.095 0.270 1.557261e-12 2
## CPSF2 4.270066e-17 -0.2046448 0.050 0.190 1.562929e-12 2
## MSH2.2 4.496379e-17 0.1696997 0.044 0.179 1.645765e-12 2
## RSRP1.1 4.501168e-17 1.5541319 0.547 0.631 1.647517e-12 2
## CIAO1.1 4.522716e-17 0.4574155 0.095 0.269 1.655404e-12 2
## BEX4 4.590378e-17 -1.5373882 0.015 0.127 1.680170e-12 2
## PLAA 4.680671e-17 -0.1201310 0.056 0.201 1.713219e-12 2
## NOC4L.1 4.782198e-17 -0.5165123 0.023 0.141 1.750380e-12 2
## PRIM1.2 4.795548e-17 -0.7777475 0.017 0.130 1.755266e-12 2
## DCAF13.1 4.821755e-17 -0.1973735 0.042 0.177 1.764859e-12 2
## FBXO9.1 5.155596e-17 0.5529532 0.090 0.262 1.887051e-12 2
## CENPW.2 5.457952e-17 -2.2672155 0.005 0.106 1.997720e-12 2
## GART.1 5.473153e-17 0.2153258 0.077 0.238 2.003284e-12 2
## PTTG1IP.1 5.573997e-17 0.3264936 0.084 0.251 2.040195e-12 2
## ACADS.1 5.603224e-17 -1.0833949 0.014 0.124 2.050892e-12 2
## ENSA 5.770395e-17 0.6892108 0.256 0.569 2.112080e-12 2
## GRAMD1A.1 6.193848e-17 0.5013192 0.096 0.273 2.267072e-12 2
## NMRAL1.1 6.203358e-17 0.6063228 0.065 0.217 2.270553e-12 2
## NCOA2.1 6.424944e-17 2.2288586 0.388 0.339 2.351658e-12 2
## PRRC2A 6.569673e-17 0.2906744 0.089 0.257 2.404632e-12 2
## FAHD2B 6.593254e-17 -1.5165619 0.011 0.118 2.413263e-12 2
## ESYT1.2 6.613563e-17 0.4750943 0.164 0.390 2.420696e-12 2
## DNTTIP1 6.684906e-17 -0.1242818 0.033 0.160 2.446809e-12 2
## MRPL49 6.866111e-17 -0.5675211 0.023 0.140 2.513134e-12 2
## OAS2 6.905146e-17 0.2379981 0.072 0.228 2.527421e-12 2
## SLC4A1AP 6.997584e-17 0.2854232 0.059 0.205 2.561256e-12 2
## WRAP73 7.263077e-17 -0.3940236 0.029 0.152 2.658432e-12 2
## SFPQ.1 7.467052e-17 0.3404097 0.299 0.635 2.733090e-12 2
## HIST3H2A.2 7.647908e-17 0.1742857 0.056 0.199 2.799287e-12 2
## ILKAP.1 8.037969e-17 0.3953672 0.075 0.235 2.942057e-12 2
## A1BG.1 8.081659e-17 -1.9991754 0.012 0.120 2.958049e-12 2
## ADPRHL2 8.190523e-17 -0.1880007 0.038 0.167 2.997895e-12 2
## ASB8 8.208792e-17 -0.1474906 0.042 0.175 3.004582e-12 2
## PCIF1.1 8.658465e-17 -0.1045468 0.044 0.178 3.169172e-12 2
## EBLN3P 8.714875e-17 0.1403616 0.068 0.220 3.189819e-12 2
## ZBTB7A.1 8.735891e-17 0.5367005 0.137 0.341 3.197511e-12 2
## LEPROT 8.795761e-17 -0.7467600 0.024 0.142 3.219424e-12 2
## RRS1.1 8.811726e-17 -0.2528881 0.030 0.154 3.225268e-12 2
## GTF2H3.1 8.867958e-17 0.2479114 0.048 0.186 3.245850e-12 2
## PRKRA 8.876901e-17 -0.3577555 0.041 0.172 3.249123e-12 2
## TAPBP.1 8.958465e-17 0.3032113 0.274 0.597 3.278977e-12 2
## ERP44 9.342461e-17 0.5739226 0.168 0.401 3.419528e-12 2
## SLC25A22 9.410100e-17 -2.2681191 0.008 0.111 3.444285e-12 2
## SCRN2 9.644936e-17 -0.5716460 0.036 0.163 3.530240e-12 2
## CXorf40B.1 9.859634e-17 -0.5159872 0.035 0.161 3.608823e-12 2
## CHP1 1.015351e-16 -0.2318478 0.033 0.158 3.716386e-12 2
## SAP30 1.033671e-16 -0.3543072 0.030 0.153 3.783444e-12 2
## BDH2 1.034914e-16 -0.7494314 0.024 0.142 3.787994e-12 2
## NCAPD2.2 1.129170e-16 -0.2140849 0.050 0.187 4.132987e-12 2
## PSMA1 1.130848e-16 0.3509299 0.305 0.652 4.139129e-12 2
## TP53.2 1.160208e-16 -0.1987760 0.030 0.153 4.246593e-12 2
## FAM98C.1 1.185499e-16 -0.6741145 0.024 0.141 4.339163e-12 2
## ELK1 1.188086e-16 -0.4086147 0.021 0.136 4.348633e-12 2
## AARS.2 1.204938e-16 0.2680252 0.057 0.200 4.410314e-12 2
## PDRG1 1.232854e-16 -1.6864579 0.008 0.110 4.512493e-12 2
## CARD19.1 1.273028e-16 -0.1282256 0.033 0.158 4.659538e-12 2
## KIF20B.2 1.280154e-16 -0.2861455 0.054 0.194 4.685618e-12 2
## OARD1 1.307187e-16 -0.3954243 0.035 0.161 4.784567e-12 2
## CFAP20.1 1.314739e-16 -0.1622266 0.047 0.182 4.812209e-12 2
## FAM110A 1.365223e-16 -0.9177134 0.015 0.124 4.996987e-12 2
## SEC22B.1 1.370459e-16 0.2455795 0.077 0.235 5.016155e-12 2
## HCP5 1.392032e-16 0.1255242 0.071 0.224 5.095115e-12 2
## NSL1 1.422027e-16 0.1277971 0.074 0.229 5.204904e-12 2
## CD82 1.425160e-16 0.1475104 0.128 0.322 5.216371e-12 2
## RAB21 1.452966e-16 0.1929883 0.071 0.224 5.318145e-12 2
## ACADVL.1 1.463288e-16 0.5278843 0.162 0.389 5.355927e-12 2
## MRPL19 1.465247e-16 -0.2959153 0.033 0.158 5.363098e-12 2
## LPCAT4.1 1.483374e-16 0.2835054 0.069 0.221 5.429447e-12 2
## MSH6.2 1.525672e-16 -0.2775529 0.036 0.163 5.584264e-12 2
## COPRS.2 1.542992e-16 -2.5335732 0.005 0.103 5.647658e-12 2
## ZDHHC4 1.551075e-16 -1.7883519 0.005 0.103 5.677245e-12 2
## ARFGAP2 1.556188e-16 0.2170304 0.066 0.216 5.695960e-12 2
## DMAC2 1.585268e-16 -1.6139477 0.011 0.115 5.802396e-12 2
## UTP14A 1.617120e-16 -0.5846000 0.023 0.138 5.918983e-12 2
## CSNK1G2 1.636275e-16 0.6452149 0.122 0.314 5.989093e-12 2
## HIST1H1D.2 1.644574e-16 0.1947468 0.192 0.425 6.019471e-12 2
## C8orf76 1.653697e-16 -0.3992940 0.027 0.146 6.052862e-12 2
## R3HCC1 1.662096e-16 -0.1014082 0.032 0.155 6.083605e-12 2
## TOR1AIP1.1 1.683444e-16 0.4218240 0.123 0.317 6.161742e-12 2
## TM9SF1.1 1.700402e-16 -0.3313176 0.027 0.146 6.223812e-12 2
## RRP1B 1.749205e-16 0.4353349 0.111 0.295 6.402439e-12 2
## PRCC 1.761889e-16 -0.4001724 0.038 0.165 6.448866e-12 2
## ASF1B.2 1.882219e-16 -0.8760221 0.021 0.135 6.889299e-12 2
## SART1 1.957195e-16 0.2571959 0.059 0.202 7.163724e-12 2
## XPA 1.983040e-16 0.2718538 0.048 0.184 7.258323e-12 2
## TRPV2.2 2.008912e-16 0.3491572 0.104 0.282 7.353020e-12 2
## TMEM218.1 2.009415e-16 -2.1306352 0.008 0.109 7.354863e-12 2
## NOP14.1 2.056120e-16 0.1616005 0.096 0.266 7.525809e-12 2
## CENPH.2 2.063033e-16 -0.6296554 0.015 0.123 7.551112e-12 2
## CDC37 2.104028e-16 0.5235132 0.293 0.637 7.701162e-12 2
## ERGIC2 2.130492e-16 0.2739165 0.081 0.242 7.798027e-12 2
## DYNLL2 2.140780e-16 -0.1883180 0.044 0.176 7.835683e-12 2
## PIP4P1 2.171198e-16 -0.2285265 0.036 0.162 7.947018e-12 2
## TBRG1.1 2.194489e-16 0.3384821 0.110 0.292 8.032268e-12 2
## PIK3IP1.1 2.237991e-16 0.2042936 0.105 0.280 8.191495e-12 2
## CHMP3 2.304693e-16 0.4367191 0.072 0.226 8.435636e-12 2
## PARVB.1 2.336625e-16 0.4372976 0.071 0.223 8.552514e-12 2
## DESI1.1 2.451876e-16 0.1346335 0.053 0.191 8.974357e-12 2
## SMAP2.1 2.479482e-16 0.3271092 0.263 0.571 9.075400e-12 2
## DDX46 2.530314e-16 0.7253612 0.205 0.465 9.261456e-12 2
## C12orf45.1 2.639910e-16 -1.5510621 0.006 0.105 9.662598e-12 2
## THEM4.2 2.664448e-16 0.3174553 0.069 0.219 9.752411e-12 2
## TK1.2 2.678937e-16 -2.7487363 0.008 0.108 9.805444e-12 2
## PLSCR1 2.747000e-16 -0.7512143 0.029 0.148 1.005457e-11 2
## C12orf65 2.768312e-16 0.1430421 0.060 0.203 1.013258e-11 2
## RBM23.1 2.776668e-16 0.3072940 0.059 0.201 1.016316e-11 2
## DRAM2 2.842961e-16 -0.3136829 0.041 0.169 1.040580e-11 2
## SRPK1.1 3.000503e-16 0.4440355 0.120 0.310 1.098244e-11 2
## TAPBPL 3.122525e-16 -0.1797384 0.050 0.185 1.142907e-11 2
## FLII 3.248733e-16 0.3490663 0.096 0.265 1.189101e-11 2
## GFI1.1 3.306631e-16 0.1092711 0.069 0.219 1.210293e-11 2
## TM2D2 3.364333e-16 -1.0540115 0.018 0.128 1.231413e-11 2
## DDX42 3.476195e-16 0.2590153 0.098 0.268 1.272357e-11 2
## HMCES 3.538739e-16 0.3062526 0.090 0.255 1.295249e-11 2
## LPGAT1 3.587500e-16 0.5893526 0.122 0.311 1.313097e-11 2
## SWI5 3.797788e-16 -0.5248345 0.020 0.131 1.390066e-11 2
## ABTB1.2 3.867453e-16 0.1962551 0.047 0.180 1.415565e-11 2
## CNOT11 4.035529e-16 0.4402010 0.083 0.243 1.477084e-11 2
## JAML.1 4.082371e-16 0.4466158 0.134 0.331 1.494229e-11 2
## ALDH18A1 4.185534e-16 -0.7195099 0.026 0.141 1.531989e-11 2
## MCM2.2 4.307790e-16 -0.6205925 0.026 0.141 1.576737e-11 2
## FBLN5 4.466423e-16 -2.2930491 0.005 0.101 1.634800e-11 2
## RAB22A.1 4.540348e-16 0.3033497 0.087 0.249 1.661858e-11 2
## TRIM27 4.580318e-16 0.2435322 0.086 0.247 1.676488e-11 2
## TARBP2 4.651750e-16 -0.2871366 0.018 0.127 1.702633e-11 2
## U2SURP 4.680563e-16 0.5923816 0.161 0.380 1.713180e-11 2
## PRAF2 4.755895e-16 -1.2760703 0.011 0.112 1.740753e-11 2
## TRUB2 4.902003e-16 -1.2943851 0.020 0.129 1.794231e-11 2
## MPI.1 5.222166e-16 -0.5468878 0.030 0.148 1.911417e-11 2
## EIF5B 5.440191e-16 0.5258710 0.152 0.362 1.991219e-11 2
## SEPHS1.1 5.666984e-16 -0.1432094 0.030 0.148 2.074229e-11 2
## ABHD16A 5.712849e-16 -0.1202177 0.044 0.173 2.091017e-11 2
## GMIP.1 5.751679e-16 0.4574800 0.096 0.265 2.105230e-11 2
## CSE1L.2 5.768587e-16 -0.1130391 0.041 0.167 2.111418e-11 2
## TCERG1.1 5.920005e-16 0.5891357 0.134 0.332 2.166840e-11 2
## MDP1 5.992081e-16 -0.6006551 0.023 0.135 2.193222e-11 2
## MAPK3.1 6.017306e-16 -1.4628313 0.014 0.118 2.202454e-11 2
## KIAA2013.1 6.030879e-16 -0.3598615 0.045 0.176 2.207422e-11 2
## GOPC.1 6.032460e-16 0.4271435 0.101 0.272 2.208001e-11 2
## IARS.2 6.071840e-16 0.5548185 0.111 0.290 2.222415e-11 2
## DDX23.1 6.166591e-16 0.2564596 0.057 0.196 2.257096e-11 2
## RPUSD1.1 6.226774e-16 -0.1711888 0.029 0.146 2.279124e-11 2
## FAHD2A 6.251270e-16 -0.9721991 0.014 0.118 2.288090e-11 2
## ITGAL.1 6.343917e-16 1.6340336 0.525 0.609 2.322001e-11 2
## TIMM29 6.348616e-16 -1.0988864 0.012 0.115 2.323720e-11 2
## PGM2 6.533394e-16 -0.2373853 0.035 0.156 2.391353e-11 2
## TMF1.2 6.574506e-16 0.4636484 0.176 0.408 2.406401e-11 2
## C16orf91 6.631730e-16 -0.7445503 0.012 0.115 2.427346e-11 2
## RBBP8.2 6.660555e-16 -0.3116425 0.032 0.151 2.437896e-11 2
## RETREG2.1 6.671564e-16 0.2234647 0.062 0.204 2.441926e-11 2
## DOHH 6.687774e-16 -1.6352930 0.011 0.112 2.447859e-11 2
## LRPAP1.1 6.692072e-16 0.5565727 0.134 0.330 2.449432e-11 2
## RPP38 6.711030e-16 -0.5826049 0.018 0.126 2.456371e-11 2
## RRAS 6.845358e-16 -0.9821672 0.020 0.129 2.505538e-11 2
## ARL8B 6.886622e-16 0.2157429 0.087 0.249 2.520641e-11 2
## LINC00892.2 7.186168e-16 -0.2649626 0.047 0.177 2.630281e-11 2
## DNAJA3 7.278098e-16 -0.3430045 0.026 0.140 2.663929e-11 2
## ORAI3.1 7.459708e-16 -0.3858160 0.033 0.153 2.730402e-11 2
## NELFB.1 7.545492e-16 0.2056557 0.068 0.214 2.761801e-11 2
## INTS10 7.546142e-16 0.3313447 0.075 0.227 2.762039e-11 2
## CIR1 7.591273e-16 0.1470050 0.074 0.225 2.778558e-11 2
## YIPF2 7.904454e-16 -1.7855915 0.009 0.108 2.893188e-11 2
## GPR65.1 8.075521e-16 0.2720514 0.080 0.234 2.955802e-11 2
## MIA3 8.497803e-16 0.4311910 0.131 0.324 3.110366e-11 2
## PTPRCAP.1 8.534412e-16 -1.3702581 0.018 0.125 3.123765e-11 2
## PSMD14.1 8.910081e-16 0.5537356 0.126 0.317 3.261268e-11 2
## RABGGTA 9.041475e-16 -0.2006718 0.026 0.139 3.309361e-11 2
## HPF1.1 9.127457e-16 -0.6525785 0.023 0.134 3.340832e-11 2
## BCAP29.1 9.951289e-16 0.1947692 0.066 0.211 3.642371e-11 2
## TRIM21 9.980869e-16 -0.4604921 0.032 0.149 3.653198e-11 2
## MFSD5 1.006006e-15 -0.4773613 0.041 0.165 3.682185e-11 2
## FAM3A 1.015005e-15 -0.1737799 0.024 0.136 3.715122e-11 2
## IRF2.1 1.027970e-15 0.3692209 0.111 0.288 3.762576e-11 2
## CSTF1 1.059685e-15 -0.1077587 0.030 0.147 3.878659e-11 2
## SMIM30 1.098414e-15 -1.8324558 0.005 0.099 4.020414e-11 2
## BCKDK 1.112223e-15 -0.9358388 0.017 0.122 4.070959e-11 2
## RPF1 1.113361e-15 -0.3361336 0.035 0.155 4.075123e-11 2
## BATF.1 1.143171e-15 0.6756103 0.128 0.317 4.184236e-11 2
## PPHLN1 1.149186e-15 0.5532516 0.156 0.370 4.206252e-11 2
## HSPA13 1.153937e-15 -0.4725533 0.044 0.171 4.223640e-11 2
## ING4.1 1.167789e-15 -0.5336355 0.024 0.136 4.274341e-11 2
## RTL8C.1 1.188746e-15 -0.6783743 0.023 0.133 4.351050e-11 2
## RBBP6 1.198681e-15 0.6639004 0.159 0.372 4.387411e-11 2
## WDR82 1.203504e-15 0.4127545 0.108 0.284 4.405064e-11 2
## P4HTM 1.204514e-15 -0.1974438 0.044 0.171 4.408764e-11 2
## PPIL1.2 1.247880e-15 -0.9021156 0.008 0.104 4.567492e-11 2
## THAP1 1.261067e-15 -0.8079287 0.021 0.130 4.615759e-11 2
## ZCCHC9 1.278451e-15 -0.8902337 0.018 0.124 4.679385e-11 2
## DTD1.1 1.341631e-15 0.6321926 0.080 0.233 4.910638e-11 2
## ST6GALNAC4 1.354142e-15 -0.6117367 0.017 0.121 4.956429e-11 2
## ZFAND2A 1.389246e-15 -0.8368496 0.014 0.116 5.084920e-11 2
## NOP58 1.405157e-15 0.5892582 0.195 0.441 5.143155e-11 2
## UBN1 1.437118e-15 0.3495492 0.093 0.257 5.260139e-11 2
## ISYNA1.2 1.461746e-15 -2.3076726 0.005 0.098 5.350284e-11 2
## PGRMC1.2 1.485531e-15 -0.5385882 0.023 0.132 5.437339e-11 2
## KYAT3.1 1.550044e-15 0.1542206 0.035 0.154 5.673470e-11 2
## PARP9.2 1.572970e-15 0.2802089 0.078 0.230 5.757383e-11 2
## CD53.1 1.594555e-15 0.1349455 0.370 0.742 5.836389e-11 2
## NTHL1 1.602095e-15 -0.7138176 0.014 0.115 5.863988e-11 2
## NEDD9.1 1.614032e-15 2.0306243 0.408 0.377 5.907681e-11 2
## FLAD1 1.629013e-15 -0.1808684 0.027 0.140 5.962514e-11 2
## DGKZ.2 1.632451e-15 0.6444024 0.129 0.321 5.975097e-11 2
## MAP2K3 1.676732e-15 0.2950894 0.059 0.196 6.137176e-11 2
## DMAC2L 1.697429e-15 -0.6602482 0.024 0.135 6.212931e-11 2
## CTSH 1.700616e-15 -0.2887607 0.033 0.151 6.224595e-11 2
## GET4.1 1.765127e-15 -0.7549829 0.017 0.121 6.460717e-11 2
## NOL11.1 1.767190e-15 0.3270119 0.050 0.180 6.468270e-11 2
## STMN3.1 1.769040e-15 0.1051637 0.062 0.201 6.475039e-11 2
## GPR137 1.772630e-15 -0.2417341 0.023 0.132 6.488179e-11 2
## OCEL1 1.783985e-15 -0.2409540 0.021 0.129 6.529741e-11 2
## STARD3NL.1 1.830010e-15 -0.1462994 0.027 0.140 6.698203e-11 2
## UNC119.1 1.838190e-15 0.2280147 0.051 0.183 6.728143e-11 2
## DDX56.1 1.863759e-15 0.3804404 0.053 0.186 6.821730e-11 2
## RALB 1.869177e-15 -0.2802994 0.041 0.164 6.841561e-11 2
## HSD17B8.1 1.888694e-15 -0.9560652 0.015 0.118 6.912997e-11 2
## GALT 1.896953e-15 0.1972289 0.074 0.221 6.943227e-11 2
## PPP2R3C 1.909644e-15 0.4123124 0.056 0.191 6.989678e-11 2
## MYH9.1 1.975049e-15 1.2122541 0.583 0.729 7.229074e-11 2
## KATNBL1.1 2.009635e-15 0.2449969 0.059 0.196 7.355666e-11 2
## CRLS1 2.018499e-15 -0.1115165 0.041 0.164 7.388108e-11 2
## TPST2.2 2.099695e-15 0.4493246 0.120 0.302 7.685303e-11 2
## DGAT1 2.212119e-15 -0.1251435 0.026 0.137 8.096796e-11 2
## CNOT3 2.227619e-15 -0.9967348 0.018 0.123 8.153533e-11 2
## NOLC1.2 2.261345e-15 0.3993942 0.092 0.253 8.276975e-11 2
## SRSF4 2.314665e-15 0.4917165 0.197 0.443 8.472137e-11 2
## PKMYT1.2 2.318450e-15 -2.2157681 0.005 0.097 8.485990e-11 2
## AC100810.1 2.321223e-15 -2.6188455 0.005 0.097 8.496140e-11 2
## ARAF.1 2.331766e-15 0.3503218 0.056 0.190 8.534730e-11 2
## USE1 2.368009e-15 0.1343689 0.035 0.153 8.667387e-11 2
## ARMT1 2.383089e-15 -0.9228398 0.015 0.117 8.722582e-11 2
## SEC23B.2 2.389297e-15 0.4352637 0.074 0.221 8.745303e-11 2
## CREB3 2.392950e-15 -0.6860991 0.017 0.120 8.758676e-11 2
## CD28.1 2.397875e-15 0.5737236 0.132 0.323 8.776702e-11 2
## HENMT1.1 2.454358e-15 -0.6040553 0.024 0.134 8.983440e-11 2
## CDK9.1 2.469446e-15 0.5651608 0.080 0.231 9.038668e-11 2
## USP16 2.530803e-15 0.4320888 0.116 0.293 9.263246e-11 2
## BSCL2 2.576981e-15 -0.5123574 0.024 0.134 9.432267e-11 2
## YARS2 2.583882e-15 -0.7560225 0.023 0.131 9.457525e-11 2
## RFC1.1 2.632082e-15 0.4544035 0.116 0.293 9.633946e-11 2
## TMUB2.1 2.632853e-15 -0.2860411 0.029 0.142 9.636768e-11 2
## NFYC.1 2.639124e-15 0.4959560 0.081 0.235 9.659723e-11 2
## SLC38A5 2.642522e-15 -0.8503756 0.023 0.131 9.672158e-11 2
## BNIP3L.1 2.687891e-15 0.1360667 0.071 0.215 9.838220e-11 2
## SBDS.1 2.712034e-15 0.1534059 0.086 0.241 9.926586e-11 2
## NAP1L4 2.725674e-15 0.4028573 0.280 0.597 9.976513e-11 2
## PRKACA 2.764567e-15 -0.5382145 0.023 0.131 1.011887e-10 2
## EIF5 2.831691e-15 0.4398092 0.260 0.556 1.036455e-10 2
## APOL3 2.846858e-15 0.4409524 0.065 0.205 1.042007e-10 2
## DPH5.1 2.881034e-15 -0.5370017 0.021 0.128 1.054516e-10 2
## VPS36 2.911633e-15 0.5793199 0.119 0.299 1.065716e-10 2
## MYADM.2 2.943650e-15 0.5428296 0.156 0.365 1.077435e-10 2
## TXNRD1.1 2.969287e-15 0.5644442 0.128 0.313 1.086818e-10 2
## CCDC12.1 3.007501e-15 0.6841360 0.137 0.330 1.100805e-10 2
## DPAGT1 3.035104e-15 -1.1382874 0.021 0.128 1.110909e-10 2
## MFAP1.1 3.060518e-15 0.3496668 0.057 0.192 1.120211e-10 2
## FGD5-AS1 3.082579e-15 -0.4255602 0.057 0.191 1.128286e-10 2
## TBCC.1 3.153967e-15 0.4592395 0.072 0.218 1.154415e-10 2
## MOAP1 3.252178e-15 -1.0745144 0.017 0.119 1.190362e-10 2
## CCL28 3.319602e-15 -1.9399314 0.009 0.105 1.215041e-10 2
## SNX1 3.361773e-15 0.4949106 0.123 0.306 1.230476e-10 2
## HDGFL2.1 3.397063e-15 0.4670404 0.068 0.211 1.243393e-10 2
## NBN.1 3.452921e-15 0.3986413 0.065 0.205 1.263838e-10 2
## ZNF580.1 3.468394e-15 0.3976238 0.057 0.192 1.269502e-10 2
## SUPT7L.1 3.549688e-15 0.2220512 0.060 0.197 1.299257e-10 2
## SF3B1.1 3.568681e-15 0.4139329 0.293 0.630 1.306209e-10 2
## KAT2B.1 3.583531e-15 2.1743349 0.405 0.382 1.311644e-10 2
## ARFIP2.1 3.661424e-15 -0.3147017 0.023 0.130 1.340154e-10 2
## LYRM4.1 3.714833e-15 0.1735689 0.044 0.168 1.359703e-10 2
## CD300A.1 3.787764e-15 -0.1941453 0.039 0.160 1.386398e-10 2
## DNAJC2.2 3.791371e-15 0.2803721 0.077 0.225 1.387718e-10 2
## PARP8.1 3.873795e-15 1.7185683 0.513 0.606 1.417887e-10 2
## THOC2 3.911236e-15 0.6068927 0.147 0.349 1.431591e-10 2
## NARS 3.947837e-15 0.1949174 0.059 0.193 1.444987e-10 2
## TERF1.1 3.977407e-15 0.2514173 0.056 0.188 1.455810e-10 2
## KLHDC2 4.001767e-15 0.3398799 0.071 0.214 1.464727e-10 2
## ELOVL5 4.018645e-15 0.5542532 0.170 0.387 1.470904e-10 2
## EIF4G1 4.037463e-15 0.3118618 0.155 0.359 1.477792e-10 2
## SHC1 4.114410e-15 0.2122440 0.048 0.176 1.505956e-10 2
## ABCF1 4.121485e-15 0.3884071 0.108 0.279 1.508546e-10 2
## ZFP36L1 4.156769e-15 0.4283629 0.265 0.553 1.521461e-10 2
## SAMHD1.2 4.210181e-15 0.2894666 0.326 0.673 1.541010e-10 2
## KIAA1143 4.215406e-15 0.1760631 0.045 0.170 1.542923e-10 2
## ASPSCR1 4.248210e-15 0.1981536 0.041 0.162 1.554930e-10 2
## PES1 4.330621e-15 0.2027343 0.042 0.165 1.585094e-10 2
## ARHGAP9.1 4.392417e-15 0.6381611 0.188 0.423 1.607712e-10 2
## C12orf76 4.401835e-15 -0.5788066 0.023 0.130 1.611160e-10 2
## SNX10.1 4.495742e-15 0.1603765 0.057 0.191 1.645531e-10 2
## SDCBP 4.711697e-15 0.5939623 0.143 0.339 1.724575e-10 2
## PHGDH.2 4.891237e-15 -1.6936238 0.006 0.098 1.790291e-10 2
## IDH3A.2 4.893866e-15 -0.3663018 0.024 0.132 1.791253e-10 2
## ALG1 5.009892e-15 -0.9497187 0.023 0.129 1.833720e-10 2
## UHMK1 5.224908e-15 0.4704532 0.123 0.307 1.912421e-10 2
## FAM107B.1 5.224989e-15 0.4778920 0.238 0.510 1.912451e-10 2
## LIG1.2 5.258779e-15 0.1396971 0.045 0.169 1.924818e-10 2
## RIOK3 5.271342e-15 0.5546336 0.117 0.296 1.929416e-10 2
## NCAPG2.2 5.306630e-15 -1.4314183 0.015 0.115 1.942333e-10 2
## PRMT5.1 5.454382e-15 -1.0890209 0.014 0.112 1.996413e-10 2
## PITPNA.1 5.472410e-15 0.4575000 0.069 0.212 2.003012e-10 2
## CENPU.2 5.599842e-15 -0.6041002 0.015 0.115 2.049654e-10 2
## UBR7.1 5.665320e-15 -0.4372558 0.027 0.137 2.073620e-10 2
## PRPF40A.1 5.701570e-15 0.5189792 0.206 0.455 2.086889e-10 2
## MRPL23.1 5.761274e-15 -0.4781389 0.012 0.109 2.108741e-10 2
## ISCA1 5.820788e-15 0.7303723 0.093 0.253 2.130525e-10 2
## FOXN3 5.858625e-15 1.9474012 0.471 0.517 2.144374e-10 2
## KTN1 5.960881e-15 0.4966974 0.167 0.383 2.181802e-10 2
## CHEK1.2 6.096984e-15 -0.9734604 0.018 0.120 2.231618e-10 2
## TMEM199 6.167129e-15 -0.4049522 0.029 0.140 2.257293e-10 2
## CIZ1 6.239551e-15 -0.1345058 0.038 0.156 2.283801e-10 2
## BAZ1B.1 6.381979e-15 0.4945304 0.162 0.372 2.335932e-10 2
## G3BP2.2 6.483519e-15 0.5178941 0.239 0.519 2.373098e-10 2
## WBP11.1 6.494265e-15 0.5145869 0.107 0.276 2.377031e-10 2
## TGDS 6.572338e-15 -0.6446859 0.023 0.128 2.405607e-10 2
## SLC44A2.1 6.628299e-15 0.6803632 0.120 0.299 2.426090e-10 2
## DDX50.1 6.958205e-15 0.5865458 0.077 0.224 2.546842e-10 2
## METTL21A.1 7.001708e-15 -0.5421873 0.020 0.123 2.562765e-10 2
## PURA 7.028460e-15 0.8172739 0.128 0.314 2.572557e-10 2
## FBXL16.1 7.152128e-15 -0.6241301 0.027 0.136 2.617822e-10 2
## SAFB 7.214807e-15 0.6282573 0.119 0.297 2.640764e-10 2
## TRIM47 7.375977e-15 -0.9828018 0.018 0.120 2.699755e-10 2
## ERI3 7.378642e-15 0.2315954 0.056 0.187 2.700730e-10 2
## RAB9A.1 7.414954e-15 -0.4544320 0.018 0.120 2.714021e-10 2
## DEAF1 7.504382e-15 -0.2230974 0.033 0.147 2.746754e-10 2
## GLUD1 7.604982e-15 0.6560203 0.120 0.298 2.783575e-10 2
## BCL2A1.1 7.615967e-15 -0.2989540 0.029 0.139 2.787596e-10 2
## GPANK1 7.724891e-15 0.2069404 0.033 0.147 2.827465e-10 2
## GRWD1.1 7.725410e-15 -0.3514072 0.021 0.125 2.827654e-10 2
## LHPP 7.738426e-15 -0.1169133 0.032 0.145 2.832419e-10 2
## UHRF1.2 7.790646e-15 -0.3768875 0.021 0.125 2.851532e-10 2
## HSPH1.2 7.809095e-15 0.5209411 0.141 0.335 2.858285e-10 2
## CGAS 7.849783e-15 0.4445686 0.066 0.204 2.873178e-10 2
## GPX7 7.901625e-15 -0.6755770 0.018 0.120 2.892153e-10 2
## TM9SF3.1 7.975658e-15 0.4031536 0.128 0.311 2.919250e-10 2
## PRPSAP2 8.068707e-15 0.2010410 0.042 0.163 2.953308e-10 2
## BCAS4.2 8.200577e-15 0.1115705 0.054 0.184 3.001575e-10 2
## CRYL1 8.449356e-15 -0.6479843 0.014 0.111 3.092633e-10 2
## IFT20 8.956965e-15 -0.2378753 0.027 0.136 3.278428e-10 2
## COQ5 8.978177e-15 -0.6528957 0.018 0.119 3.286192e-10 2
## GEMIN6 9.368880e-15 -1.3057216 0.008 0.099 3.429197e-10 2
## FAM111A.1 9.432757e-15 0.1744029 0.098 0.257 3.452578e-10 2
## CPSF4.1 9.455886e-15 -0.6543353 0.018 0.119 3.461044e-10 2
## TSPAN3.1 9.584854e-15 -0.6941105 0.017 0.116 3.508248e-10 2
## ECSIT 9.695937e-15 -0.4356945 0.023 0.127 3.548907e-10 2
## OSTM1.1 9.700089e-15 0.3008613 0.077 0.222 3.550426e-10 2
## TEN1 9.779692e-15 -0.9858221 0.017 0.116 3.579563e-10 2
## GLT8D1 9.870440e-15 -0.2455931 0.041 0.160 3.612778e-10 2
## VHL 9.924551e-15 0.1810741 0.062 0.196 3.632584e-10 2
## PIGH 1.019405e-14 -0.9276264 0.017 0.116 3.731225e-10 2
## UTP6 1.050732e-14 0.1328544 0.048 0.173 3.845889e-10 2
## LGALS9.2 1.056883e-14 0.2523698 0.039 0.157 3.868403e-10 2
## C1orf174 1.070341e-14 -0.8526793 0.020 0.122 3.917662e-10 2
## CASP8AP2.1 1.092087e-14 0.3765431 0.072 0.214 3.997257e-10 2
## GSTM1.1 1.106087e-14 -3.4146252 0.003 0.090 4.048499e-10 2
## RBM18.1 1.200369e-14 0.3754518 0.050 0.174 4.393590e-10 2
## FAS.1 1.218183e-14 0.3512820 0.105 0.271 4.458794e-10 2
## MIIP.1 1.233884e-14 0.5597011 0.069 0.208 4.516261e-10 2
## MYO1G.2 1.256209e-14 0.5585126 0.239 0.510 4.597977e-10 2
## DLST.1 1.256499e-14 0.4831368 0.062 0.196 4.599039e-10 2
## BCL11B 1.296119e-14 1.7733596 0.490 0.555 4.744056e-10 2
## TMEM138 1.324128e-14 -0.2666524 0.021 0.124 4.846573e-10 2
## CHTF8.1 1.375742e-14 -1.1092719 0.015 0.113 5.035491e-10 2
## MARCH2.1 1.463872e-14 -0.1640922 0.029 0.137 5.358065e-10 2
## CYSLTR1.2 1.492324e-14 -0.2246520 0.036 0.150 5.462206e-10 2
## NPM3.2 1.514817e-14 -1.2974947 0.005 0.092 5.544532e-10 2
## STOM.2 1.598059e-14 0.3071083 0.090 0.243 5.849217e-10 2
## CHMP7 1.601602e-14 0.4574781 0.084 0.234 5.862182e-10 2
## DHX29 1.622488e-14 0.2136511 0.059 0.190 5.938630e-10 2
## MAGED1 1.625372e-14 0.3311312 0.087 0.240 5.949187e-10 2
## LPXN.1 1.626889e-14 0.4622899 0.302 0.632 5.954740e-10 2
## TUBA1A.1 1.634146e-14 0.7381416 0.254 0.539 5.981301e-10 2
## SNRNP27 1.642052e-14 -0.4746811 0.023 0.126 6.010239e-10 2
## CSTF2T.1 1.651956e-14 -0.2987884 0.027 0.134 6.046488e-10 2
## KIAA1191.1 1.674269e-14 0.2191247 0.044 0.163 6.128158e-10 2
## RBM7 1.679630e-14 -0.5336020 0.023 0.126 6.147783e-10 2
## SPOUT1 1.680881e-14 -0.5491567 0.024 0.129 6.152360e-10 2
## TNFSF14.2 1.682705e-14 0.2764253 0.077 0.220 6.159038e-10 2
## IFT27.1 1.690976e-14 0.1051728 0.026 0.132 6.189311e-10 2
## TPX2.2 1.713942e-14 -0.5226196 0.036 0.149 6.273371e-10 2
## SGO1.2 1.741699e-14 -1.7562310 0.005 0.091 6.374965e-10 2
## UTP18.1 1.758186e-14 0.3878547 0.069 0.208 6.435311e-10 2
## PDK2 1.767263e-14 -0.5933360 0.015 0.112 6.468536e-10 2
## IFI16.1 1.771539e-14 0.4637272 0.250 0.527 6.484185e-10 2
## ARV1 1.775938e-14 -0.7651591 0.014 0.109 6.500289e-10 2
## WSB2.1 1.804876e-14 -0.3280496 0.024 0.128 6.606208e-10 2
## SLC39A4.2 1.852803e-14 -0.7291176 0.014 0.109 6.781630e-10 2
## RNF214.1 1.988037e-14 0.3663550 0.075 0.217 7.276614e-10 2
## EIF2D 2.015749e-14 0.2944119 0.057 0.187 7.378043e-10 2
## GBP5.2 2.082960e-14 0.3799420 0.308 0.617 7.624049e-10 2
## TGS1 2.104260e-14 0.4140222 0.083 0.229 7.702011e-10 2
## TMEM128 2.118154e-14 -0.8636016 0.017 0.114 7.752868e-10 2
## SNUPN.1 2.138446e-14 -0.1275336 0.024 0.128 7.827140e-10 2
## FANCI.2 2.143468e-14 -0.1564555 0.044 0.161 7.845522e-10 2
## FAHD1 2.163151e-14 -2.1234189 0.003 0.088 7.917565e-10 2
## ALAS1.1 2.263727e-14 0.4093486 0.075 0.216 8.285695e-10 2
## XAF1.1 2.289351e-14 0.5007415 0.080 0.225 8.379482e-10 2
## MPHOSPH6.1 2.366567e-14 -0.1126828 0.032 0.142 8.662108e-10 2
## TRIM59 2.393897e-14 0.3280359 0.069 0.207 8.762142e-10 2
## BNIP3 2.404662e-14 0.3196323 0.042 0.160 8.801543e-10 2
## CCNK 2.501348e-14 0.1804596 0.087 0.236 9.155432e-10 2
## JMJD6 2.532047e-14 0.2128503 0.042 0.160 9.267798e-10 2
## NUP42.1 2.543821e-14 0.4107606 0.066 0.202 9.310895e-10 2
## APEX2 2.642442e-14 -0.6545075 0.017 0.114 9.671867e-10 2
## RSRC2 2.650646e-14 0.5326618 0.183 0.405 9.701895e-10 2
## GCSH.1 2.712127e-14 -1.2527697 0.006 0.093 9.926927e-10 2
## DDX41 2.720486e-14 -0.1850045 0.030 0.138 9.957523e-10 2
## STARD3.1 2.733747e-14 0.3949208 0.063 0.196 1.000606e-09 2
## VPS26A 2.753086e-14 0.5704743 0.089 0.241 1.007685e-09 2
## ATG4B.1 2.777557e-14 0.4582060 0.083 0.230 1.016641e-09 2
## LMNB2.2 2.809964e-14 -0.4273475 0.021 0.122 1.028503e-09 2
## PRKAB1 2.830097e-14 -1.1069827 0.018 0.116 1.035872e-09 2
## DUSP12 2.864379e-14 -0.1695283 0.026 0.130 1.048420e-09 2
## OIP5-AS1 2.915833e-14 0.4038734 0.147 0.343 1.067253e-09 2
## GTPBP8.1 2.955443e-14 -0.3301614 0.023 0.124 1.081751e-09 2
## RNPEP 2.963832e-14 0.1491869 0.048 0.170 1.084822e-09 2
## SF3B3.1 3.030610e-14 0.3976890 0.092 0.245 1.109264e-09 2
## PPP5C.1 3.062394e-14 0.7058541 0.084 0.233 1.120897e-09 2
## GTF2H1 3.083788e-14 0.1026475 0.050 0.172 1.128728e-09 2
## OSBP 3.085281e-14 0.3629965 0.086 0.233 1.129274e-09 2
## CCNA2.2 3.098185e-14 -3.4733238 0.005 0.090 1.133998e-09 2
## GNL2.1 3.102497e-14 0.2188767 0.051 0.175 1.135576e-09 2
## PELP1.1 3.189986e-14 0.4530290 0.053 0.178 1.167599e-09 2
## TNFRSF25.1 3.246412e-14 0.5286455 0.083 0.229 1.188252e-09 2
## STK26.1 3.273807e-14 0.3854003 0.062 0.193 1.198279e-09 2
## C6orf89 3.310602e-14 0.3318704 0.078 0.221 1.211747e-09 2
## STAT5A.1 3.326687e-14 0.1955448 0.063 0.195 1.217634e-09 2
## MSRB2.2 3.331408e-14 -2.3341762 0.006 0.093 1.219362e-09 2
## TIMM22 3.351061e-14 -0.1413752 0.024 0.127 1.226555e-09 2
## CD4.2 3.570872e-14 0.1751471 0.083 0.227 1.307011e-09 2
## CEBPZ.1 3.592138e-14 0.5831492 0.078 0.221 1.314794e-09 2
## MSRB1.1 3.660249e-14 -2.2968371 0.003 0.087 1.339724e-09 2
## ACLY.1 3.665091e-14 0.5719451 0.087 0.237 1.341496e-09 2
## SPINT2.2 3.687820e-14 -2.6723354 0.006 0.092 1.349816e-09 2
## SLC27A5 3.694715e-14 -1.0742776 0.012 0.104 1.352340e-09 2
## LPP.1 3.760796e-14 2.1473233 0.400 0.383 1.376527e-09 2
## TUBA1C.1 3.851987e-14 0.7868083 0.149 0.342 1.409904e-09 2
## ADAM15.1 3.915139e-14 -0.5627682 0.020 0.118 1.433019e-09 2
## PARD6A 3.933238e-14 -0.9400979 0.011 0.101 1.439644e-09 2
## STK10.1 3.957052e-14 1.7260399 0.484 0.547 1.448360e-09 2
## TMEM189.1 3.964598e-14 -0.1201286 0.033 0.143 1.451122e-09 2
## MYBL2.2 4.094935e-14 -1.6918670 0.008 0.095 1.498828e-09 2
## BBC3.2 4.122510e-14 0.1450760 0.072 0.208 1.508921e-09 2
## TBRG4 4.289864e-14 0.3710877 0.051 0.174 1.570176e-09 2
## LRIF1 4.432705e-14 0.3793195 0.048 0.168 1.622459e-09 2
## EEF1AKNMT 4.527759e-14 -1.1710831 0.017 0.112 1.657250e-09 2
## KPNA2.1 4.532962e-14 0.3056108 0.081 0.224 1.659155e-09 2
## HDLBP 4.596078e-14 0.5635903 0.171 0.383 1.682256e-09 2
## ARMCX3 4.669300e-14 -0.1985508 0.051 0.173 1.709057e-09 2
## EXOC5 4.712825e-14 0.1567953 0.062 0.191 1.724988e-09 2
## GRINA 4.761400e-14 -0.7560281 0.020 0.118 1.742768e-09 2
## C9orf40.2 4.802162e-14 -0.4862797 0.014 0.107 1.757687e-09 2
## OTULINL.1 5.003030e-14 0.4193039 0.072 0.209 1.831209e-09 2
## TRABD2A.1 5.122410e-14 0.6807371 0.072 0.210 1.874905e-09 2
## SGSM3 5.137898e-14 0.5445247 0.068 0.201 1.880574e-09 2
## RASA4 5.164656e-14 -0.6828168 0.021 0.120 1.890368e-09 2
## MUL1 5.289974e-14 -1.2493388 0.014 0.106 1.936236e-09 2
## NT5C3B.1 5.359079e-14 -0.7194579 0.014 0.106 1.961530e-09 2
## DDX27 5.428485e-14 0.6230353 0.071 0.207 1.986934e-09 2
## SLC16A3.1 5.491135e-14 0.1985596 0.054 0.178 2.009865e-09 2
## BARD1.2 5.518366e-14 -0.5271938 0.027 0.131 2.019832e-09 2
## OAT.2 5.667444e-14 0.3796092 0.054 0.178 2.074398e-09 2
## MED6.1 5.807480e-14 0.1912683 0.045 0.163 2.125654e-09 2
## ZMPSTE24 5.873638e-14 0.2367984 0.065 0.196 2.149869e-09 2
## KLRK1.2 5.929853e-14 0.2045176 0.200 0.414 2.170445e-09 2
## UBE2C.2 5.989576e-14 -2.4972586 0.005 0.088 2.192304e-09 2
## C6orf136 5.995297e-14 -0.4892690 0.018 0.114 2.194399e-09 2
## COQ9 6.039058e-14 -0.3068335 0.026 0.128 2.210416e-09 2
## MGME1.1 6.101236e-14 0.4444152 0.048 0.168 2.233174e-09 2
## ATP1A1 6.150928e-14 0.3993090 0.131 0.309 2.251363e-09 2
## AC093323.1 6.166526e-14 -0.2345419 0.027 0.131 2.257072e-09 2
## RINL 6.231794e-14 0.1237794 0.048 0.168 2.280961e-09 2
## FAM111B.2 6.237028e-14 -2.0402231 0.006 0.091 2.282877e-09 2
## DUSP11.1 6.379980e-14 0.2293376 0.053 0.176 2.335200e-09 2
## DIAPH3.2 6.531038e-14 -1.7664850 0.015 0.108 2.390490e-09 2
## XAB2 6.582972e-14 0.3718130 0.041 0.155 2.409499e-09 2
## AL441992.1 7.008783e-14 -3.5376304 0.003 0.085 2.565355e-09 2
## PGAP2.2 7.255688e-14 -0.5332902 0.017 0.111 2.655727e-09 2
## RIOK2 7.336172e-14 -0.5214825 0.021 0.119 2.685186e-09 2
## TPR 7.420453e-14 0.5297667 0.287 0.597 2.716034e-09 2
## TRMT6 7.504919e-14 -1.3162542 0.011 0.099 2.746950e-09 2
## SMIM27 7.608585e-14 -1.4104539 0.009 0.097 2.784894e-09 2
## RRN3 7.656551e-14 -0.3075656 0.042 0.156 2.802451e-09 2
## JKAMP.1 7.671996e-14 0.3654880 0.050 0.170 2.808104e-09 2
## CTR9.1 7.677911e-14 0.6864389 0.125 0.299 2.810269e-09 2
## CBX5.2 7.740697e-14 0.4531328 0.077 0.216 2.833250e-09 2
## ZFP91.1 7.781322e-14 0.7361146 0.093 0.244 2.848120e-09 2
## CHRNB1 7.786726e-14 -1.2610336 0.011 0.099 2.850098e-09 2
## ALG8.2 7.792501e-14 0.1139020 0.041 0.154 2.852211e-09 2
## TMEM259.2 7.846627e-14 0.7006456 0.137 0.321 2.872022e-09 2
## DHRS3.1 7.887166e-14 0.3755716 0.057 0.183 2.886861e-09 2
## ZMYND19.2 8.045979e-14 -0.3628374 0.023 0.122 2.944989e-09 2
## ZNF207 8.049831e-14 0.5841233 0.183 0.404 2.946399e-09 2
## DPP3.1 8.088545e-14 0.3916600 0.035 0.144 2.960569e-09 2
## RDH14 8.122386e-14 -0.4218531 0.021 0.119 2.972956e-09 2
## MVD 8.128587e-14 0.3015762 0.048 0.166 2.975225e-09 2
## LAPTM4B.1 8.228003e-14 -0.8270511 0.018 0.113 3.011614e-09 2
## MLH1.1 8.277621e-14 0.3770478 0.050 0.170 3.029775e-09 2
## UBA5 8.306880e-14 0.1123196 0.056 0.180 3.040484e-09 2
## COPS7B 8.322031e-14 -0.7881008 0.017 0.111 3.046030e-09 2
## DAP.1 8.784898e-14 0.2959394 0.060 0.187 3.215448e-09 2
## OGFOD1 8.799939e-14 -0.1713345 0.029 0.132 3.220954e-09 2
## PDE7A.1 8.846323e-14 1.8521040 0.480 0.549 3.237931e-09 2
## KBTBD4 9.092916e-14 -1.0309397 0.018 0.113 3.328189e-09 2
## CHPF.1 9.168628e-14 -0.9987373 0.020 0.116 3.355901e-09 2
## PITPNA-AS1 9.243868e-14 -1.1337368 0.009 0.096 3.383440e-09 2
## IFI27L1.2 9.614810e-14 -0.3734239 0.017 0.110 3.519213e-09 2
## C2orf68 9.686562e-14 -1.5976524 0.011 0.099 3.545475e-09 2
## POLR3C 9.893387e-14 -0.1331972 0.029 0.132 3.621177e-09 2
## C2orf74 9.935268e-14 -0.6200587 0.018 0.113 3.636507e-09 2
## RFX5 1.002430e-13 -0.3568534 0.024 0.124 3.669096e-09 2
## MAPK9.1 1.005458e-13 -0.2295592 0.038 0.148 3.680177e-09 2
## ILVBL.1 1.026979e-13 -0.7365870 0.015 0.107 3.758948e-09 2
## PRPF8 1.045563e-13 0.4705513 0.180 0.396 3.826971e-09 2
## LYRM1 1.058426e-13 0.2842049 0.036 0.146 3.874053e-09 2
## PPM1K 1.089476e-13 0.5820186 0.102 0.260 3.987698e-09 2
## AIDA 1.093994e-13 0.1657832 0.050 0.168 4.004237e-09 2
## PAQR4.2 1.111439e-13 -1.5863039 0.005 0.087 4.068087e-09 2
## PSMD1.1 1.137802e-13 0.5746823 0.138 0.321 4.164582e-09 2
## ACTR1B 1.143095e-13 0.5342991 0.059 0.184 4.183957e-09 2
## YBEY 1.157816e-13 -1.4406523 0.008 0.093 4.237837e-09 2
## SNHG3 1.171505e-13 -0.1863529 0.033 0.140 4.287943e-09 2
## SGO2.2 1.172394e-13 -1.3718863 0.006 0.090 4.291197e-09 2
## TMOD3.1 1.191673e-13 0.4722010 0.189 0.412 4.361760e-09 2
## ULK3 1.256367e-13 0.5286914 0.050 0.169 4.598556e-09 2
## ISOC1.1 1.283246e-13 -0.1670274 0.027 0.129 4.696936e-09 2
## CYREN 1.286252e-13 -0.6720221 0.021 0.118 4.707940e-09 2
## TUBGCP2.1 1.302855e-13 0.4525966 0.089 0.234 4.768710e-09 2
## SBNO1.1 1.307443e-13 0.4979047 0.104 0.261 4.785505e-09 2
## HIST1H1B.2 1.317399e-13 -0.8320799 0.062 0.183 4.821945e-09 2
## PIGBOS1.1 1.324578e-13 -0.2234280 0.020 0.115 4.848222e-09 2
## C20orf204 1.377818e-13 0.1269715 0.021 0.118 5.043091e-09 2
## GLUL.1 1.384280e-13 -0.5186726 0.023 0.120 5.066740e-09 2
## TOP2A.2 1.405822e-13 -1.0671503 0.021 0.117 5.145588e-09 2
## ASMTL 1.474735e-13 0.6533879 0.077 0.215 5.397825e-09 2
## MT1F.1 1.487277e-13 -1.1540964 0.009 0.095 5.443732e-09 2
## UQCRHL 1.488607e-13 -3.0794659 0.003 0.083 5.448598e-09 2
## RABEPK.1 1.503700e-13 0.1694154 0.023 0.120 5.503841e-09 2
## SLC43A3.2 1.523265e-13 -0.4700905 0.012 0.101 5.575455e-09 2
## TAF9B.1 1.538927e-13 -0.4333124 0.014 0.103 5.632779e-09 2
## TDP2.1 1.590902e-13 0.4550624 0.039 0.149 5.823020e-09 2
## CDC6.2 1.609016e-13 -0.6350774 0.012 0.100 5.889320e-09 2
## PRKCQ-AS1.2 1.609613e-13 0.6740409 0.212 0.451 5.891506e-09 2
## POLR1E.1 1.617131e-13 -0.4039348 0.015 0.106 5.919025e-09 2
## TBC1D10B.1 1.624678e-13 0.1904930 0.035 0.141 5.946645e-09 2
## AC022706.1.2 1.653482e-13 -1.6610584 0.008 0.092 6.052073e-09 2
## PDCL 1.656131e-13 0.1574119 0.035 0.142 6.061771e-09 2
## RRP9 1.665060e-13 -0.6401103 0.014 0.103 6.094451e-09 2
## DESI2.1 1.697161e-13 0.1361466 0.051 0.170 6.211948e-09 2
## HEIH 1.710858e-13 -1.4490900 0.006 0.089 6.262081e-09 2
## FAM117A.1 1.716419e-13 0.6485554 0.125 0.296 6.282437e-09 2
## DDAH2.1 1.733806e-13 -0.4456800 0.015 0.106 6.346077e-09 2
## LRRC42.2 1.751195e-13 -0.4055489 0.014 0.103 6.409725e-09 2
## TM9SF2.1 1.780847e-13 0.4757286 0.168 0.372 6.518257e-09 2
## TACO1 1.785023e-13 -0.3311827 0.020 0.114 6.533540e-09 2
## TIFA.2 1.788362e-13 -0.6020370 0.020 0.114 6.545764e-09 2
## STAT1.2 1.796433e-13 0.1163256 0.260 0.494 6.575302e-09 2
## GOLGA2 1.808268e-13 0.5034208 0.057 0.180 6.618621e-09 2
## DDX6.1 1.832412e-13 0.4986235 0.208 0.443 6.706994e-09 2
## PTPRJ.2 1.883514e-13 2.1568040 0.391 0.369 6.894038e-09 2
## F11R 1.915156e-13 -0.9027401 0.014 0.103 7.009855e-09 2
## PARP10.1 1.915657e-13 0.4340422 0.068 0.198 7.011688e-09 2
## DENND10 1.929390e-13 0.2931039 0.036 0.144 7.061954e-09 2
## UCKL1 1.949287e-13 0.5528491 0.050 0.167 7.134782e-09 2
## UBIAD1 1.972627e-13 -0.1506383 0.030 0.133 7.220210e-09 2
## TP53RK 2.022369e-13 -1.1272063 0.015 0.105 7.402275e-09 2
## RTN4.1 2.030656e-13 0.7145589 0.185 0.406 7.432606e-09 2
## ZNF326.1 2.048323e-13 0.3881539 0.063 0.190 7.497270e-09 2
## KRI1 2.066908e-13 0.8441493 0.098 0.250 7.565297e-09 2
## MARCKSL1.1 2.078170e-13 0.5910938 0.039 0.149 7.606517e-09 2
## NFYB.1 2.109821e-13 0.1178602 0.032 0.135 7.722366e-09 2
## CCDC47.1 2.131465e-13 0.6284525 0.107 0.265 7.801588e-09 2
## PLIN3.1 2.150545e-13 -0.2778659 0.026 0.124 7.871423e-09 2
## ABLIM1.2 2.151714e-13 0.5887006 0.171 0.375 7.875703e-09 2
## IL4R.1 2.163413e-13 0.4750621 0.114 0.274 7.918525e-09 2
## CWC25 2.239325e-13 0.4953641 0.080 0.217 8.196378e-09 2
## PDXK.1 2.241689e-13 0.7113952 0.065 0.193 8.205028e-09 2
## MCFD2.1 2.361822e-13 0.1550753 0.038 0.145 8.644743e-09 2
## MMAB.2 2.365975e-13 0.2336530 0.023 0.119 8.659941e-09 2
## B3GALT6 2.371368e-13 -0.6617935 0.026 0.124 8.679680e-09 2
## CGRRF1 2.375253e-13 -0.5060250 0.012 0.099 8.693902e-09 2
## PTPA 2.384076e-13 0.1099644 0.039 0.148 8.726195e-09 2
## SEC22C 2.395445e-13 0.5422591 0.069 0.200 8.767807e-09 2
## SOCS2 2.399927e-13 0.2825919 0.045 0.159 8.784214e-09 2
## OAS1.1 2.442043e-13 -0.6278554 0.020 0.113 8.938364e-09 2
## TPRA1 2.465599e-13 -0.2466003 0.027 0.127 9.024587e-09 2
## MAEA 2.571622e-13 0.5203474 0.083 0.222 9.412650e-09 2
## SAAL1.1 2.606652e-13 -0.7091511 0.011 0.096 9.540868e-09 2
## TDG 2.661725e-13 0.4497464 0.068 0.196 9.742447e-09 2
## DBP 2.684628e-13 -0.4535132 0.027 0.126 9.826275e-09 2
## ALG2 2.688030e-13 -0.1432237 0.051 0.168 9.838726e-09 2
## NCAPG.2 2.696675e-13 -1.3992764 0.008 0.090 9.870368e-09 2
## MED19 2.731670e-13 -0.2854405 0.024 0.121 9.998457e-09 2
## FAM217B 2.757735e-13 -0.1393083 0.039 0.147 1.009386e-08 2
## PTPN7.2 2.771379e-13 0.5561927 0.203 0.432 1.014380e-08 2
## ATL3.1 2.777958e-13 0.3086808 0.065 0.192 1.016788e-08 2
## RUFY1 2.838825e-13 0.3483743 0.060 0.184 1.039067e-08 2
## TIMELESS.2 2.845022e-13 -0.4972935 0.017 0.107 1.041335e-08 2
## AHNAK.2 2.846112e-13 1.3001701 0.564 0.690 1.041734e-08 2
## MPP1.1 2.875459e-13 -0.1966985 0.033 0.137 1.052476e-08 2
## ZCCHC10 2.943943e-13 -0.5413060 0.026 0.124 1.077542e-08 2
## IER5.1 2.943990e-13 0.7264878 0.161 0.356 1.077559e-08 2
## ANKRD11 2.957736e-13 1.6703230 0.481 0.544 1.082591e-08 2
## ZNF410 2.989639e-13 0.3709140 0.050 0.166 1.094268e-08 2
## FARSB.1 2.997313e-13 0.2556159 0.056 0.176 1.097077e-08 2
## CEBPG 3.006081e-13 0.1851224 0.030 0.132 1.100286e-08 2
## PSPH 3.030152e-13 -2.1269619 0.008 0.090 1.109096e-08 2
## MRS2.1 3.037429e-13 -0.5474542 0.018 0.110 1.111760e-08 2
## KATNA1 3.039391e-13 0.5447498 0.048 0.163 1.112478e-08 2
## GINM1 3.069877e-13 0.1220087 0.041 0.150 1.123636e-08 2
## CDC45.2 3.121038e-13 -1.2697309 0.011 0.095 1.142362e-08 2
## UTP23.1 3.131906e-13 0.3914363 0.050 0.166 1.146340e-08 2
## MED15.1 3.233955e-13 0.5373347 0.141 0.321 1.183692e-08 2
## ABCF3 3.238129e-13 -0.4745658 0.017 0.107 1.185220e-08 2
## GLB1 3.243996e-13 0.3804456 0.075 0.210 1.187367e-08 2
## GZMB.2 3.273475e-13 -0.9979115 0.057 0.172 1.198157e-08 2
## FAM210B 3.287444e-13 -1.4886914 0.006 0.087 1.203270e-08 2
## IGSF8.1 3.401781e-13 -0.2230027 0.045 0.157 1.245120e-08 2
## KIFC1.2 3.416040e-13 -1.6331218 0.011 0.095 1.250339e-08 2
## CNOT9.1 3.435459e-13 0.4678585 0.083 0.222 1.257447e-08 2
## DCTN1 3.554099e-13 0.5546142 0.071 0.201 1.300871e-08 2
## MYNN 3.675692e-13 0.3362764 0.035 0.139 1.345377e-08 2
## DMAP1 3.799849e-13 0.2128680 0.036 0.142 1.390821e-08 2
## KANSL1-AS1.1 4.114632e-13 -0.2018342 0.024 0.120 1.506038e-08 2
## C8orf82 4.156507e-13 -0.8517490 0.015 0.104 1.521365e-08 2
## MSMO1 4.274907e-13 -0.3859454 0.027 0.125 1.564701e-08 2
## RCC1L.1 4.297261e-13 0.3864915 0.044 0.155 1.572883e-08 2
## VCP.1 4.345966e-13 0.4796249 0.256 0.527 1.590711e-08 2
## MORF4L2 4.355971e-13 0.7974896 0.144 0.327 1.594373e-08 2
## ADA2.2 4.366158e-13 0.4599565 0.095 0.241 1.598101e-08 2
## ARID1B.1 4.572818e-13 2.0795574 0.394 0.377 1.673743e-08 2
## MMGT1 4.580096e-13 -0.1571141 0.030 0.130 1.676407e-08 2
## KIAA0040 4.606097e-13 0.5031820 0.105 0.260 1.685924e-08 2
## PLEKHB2 4.738550e-13 0.3708649 0.066 0.192 1.734404e-08 2
## MRPS28.1 4.764305e-13 0.5904280 0.059 0.180 1.743831e-08 2
## KPNA3.1 4.805980e-13 0.6251305 0.078 0.213 1.759085e-08 2
## TSEN34 4.825895e-13 -0.5882660 0.023 0.117 1.766374e-08 2
## SMARCC1.1 4.878255e-13 0.5317661 0.177 0.385 1.785539e-08 2
## C8orf33.1 4.911580e-13 -0.7567975 0.008 0.089 1.797736e-08 2
## TIMM10B 4.983299e-13 0.1205816 0.036 0.141 1.823987e-08 2
## BMS1 4.983349e-13 0.5292787 0.069 0.198 1.824005e-08 2
## NMRK1 5.057441e-13 0.4884037 0.072 0.203 1.851124e-08 2
## AAGAB 5.150024e-13 0.5070537 0.090 0.233 1.885012e-08 2
## CRTC2 5.191155e-13 0.4611865 0.056 0.175 1.900067e-08 2
## RIT1 5.250397e-13 -0.3239009 0.038 0.143 1.921750e-08 2
## TUBG1.2 5.325083e-13 -1.6658911 0.008 0.089 1.949087e-08 2
## DPP4.1 5.350512e-13 0.5343562 0.230 0.478 1.958394e-08 2
## UCK1.1 5.416637e-13 -0.1410008 0.021 0.114 1.982598e-08 2
## ORC6.2 5.420154e-13 -1.0549951 0.009 0.091 1.983885e-08 2
## LGALS8 5.466272e-13 0.3090566 0.069 0.198 2.000765e-08 2
## PDE6G 5.471993e-13 -0.7833726 0.015 0.103 2.002859e-08 2
## SLC7A6OS.1 5.501135e-13 0.2546247 0.048 0.161 2.013525e-08 2
## YIPF4 5.511265e-13 0.4222471 0.093 0.239 2.017233e-08 2
## PPWD1 5.574255e-13 0.3219574 0.083 0.220 2.040289e-08 2
## THG1L 5.616890e-13 -0.5282699 0.014 0.100 2.055894e-08 2
## MTMR14 5.638994e-13 0.6574493 0.072 0.202 2.063985e-08 2
## AARSD1 5.701895e-13 -0.2098724 0.032 0.133 2.087008e-08 2
## PPP2R5D.1 5.734056e-13 -0.1565554 0.033 0.135 2.098779e-08 2
## CCDC32 5.749863e-13 0.6893039 0.050 0.164 2.104565e-08 2
## NCAPD3.2 5.759477e-13 0.1143672 0.059 0.178 2.108084e-08 2
## WDR55 5.879256e-13 0.3374753 0.036 0.140 2.151925e-08 2
## PCYT2 5.952322e-13 0.1721391 0.033 0.135 2.178669e-08 2
## CTBS.1 6.209119e-13 0.2730822 0.072 0.202 2.272662e-08 2
## SUDS3 6.333713e-13 0.4134545 0.063 0.186 2.318266e-08 2
## AZIN1.1 6.386849e-13 0.4867638 0.075 0.207 2.337714e-08 2
## CLSPN.2 6.427222e-13 0.2619993 0.036 0.140 2.352492e-08 2
## VRK3 6.442087e-13 0.5000492 0.059 0.178 2.357933e-08 2
## CHST12.1 6.517518e-13 0.5221390 0.107 0.260 2.385542e-08 2
## NCSTN 6.531890e-13 -0.1537039 0.047 0.159 2.390802e-08 2
## ODF2.2 6.746784e-13 0.1195502 0.045 0.155 2.469458e-08 2
## POLDIP3 6.773549e-13 0.4538276 0.053 0.169 2.479255e-08 2
## TSG101.1 7.033387e-13 0.5822850 0.080 0.215 2.574360e-08 2
## SLC25A46 7.150867e-13 0.4688166 0.089 0.230 2.617360e-08 2
## RAB1A 7.202009e-13 0.5218221 0.111 0.268 2.636080e-08 2
## ZNF480 7.499138e-13 0.1052325 0.033 0.134 2.744834e-08 2
## KLHL9 7.533428e-13 -0.5493030 0.027 0.124 2.757385e-08 2
## NELFA 7.660588e-13 0.2190248 0.038 0.142 2.803928e-08 2
## CHMP4B 7.811087e-13 0.4403938 0.060 0.181 2.859014e-08 2
## LPAR2 7.835003e-13 -0.1297324 0.027 0.124 2.867768e-08 2
## TTC32 7.863338e-13 -0.2206912 0.032 0.132 2.878139e-08 2
## CTU1 8.014168e-13 -0.5021501 0.014 0.099 2.933346e-08 2
## TMEM80.1 8.017017e-13 -0.5076435 0.014 0.099 2.934388e-08 2
## BBIP1 8.068313e-13 -0.3831665 0.020 0.110 2.953164e-08 2
## DTX3L.1 8.076178e-13 0.3965748 0.075 0.205 2.956043e-08 2
## TNFRSF1B.1 8.209001e-13 0.6073969 0.165 0.359 3.004659e-08 2
## G6PC3 8.233046e-13 -1.3436781 0.008 0.087 3.013459e-08 2
## ATXN2L 8.307285e-13 0.8231711 0.144 0.325 3.040632e-08 2
## CTCF 8.352621e-13 0.6611045 0.162 0.357 3.057226e-08 2
## SHMT1.2 8.362262e-13 -0.3915711 0.023 0.115 3.060755e-08 2
## ZNF830 8.500478e-13 0.2894611 0.051 0.165 3.111345e-08 2
## TBC1D17 8.704319e-13 0.1142771 0.042 0.150 3.185955e-08 2
## ISG20L2 8.734973e-13 0.2141962 0.045 0.155 3.197175e-08 2
## RHOT2 8.855040e-13 0.6291582 0.066 0.190 3.241122e-08 2
## TNFRSF1A.1 8.912459e-13 0.5166876 0.060 0.180 3.262138e-08 2
## MED31 9.137307e-13 -0.3826159 0.024 0.118 3.344437e-08 2
## TPMT.1 9.276276e-13 -0.5963215 0.014 0.099 3.395302e-08 2
## C1orf35.2 9.295185e-13 0.7349517 0.051 0.165 3.402224e-08 2
## C12orf43.1 9.348773e-13 -0.2181923 0.029 0.126 3.421838e-08 2
## ACSL5 9.359722e-13 0.4757815 0.069 0.196 3.425846e-08 2
## TMEM216 9.425415e-13 -2.0890657 0.008 0.087 3.449890e-08 2
## RRP1 9.778772e-13 0.3290684 0.026 0.121 3.579226e-08 2
## PEMT.1 9.817995e-13 -0.5809189 0.014 0.099 3.593583e-08 2
## TRIB3 9.876892e-13 -1.4821283 0.009 0.090 3.615140e-08 2
## FUOM.1 9.904377e-13 -0.8858935 0.011 0.093 3.625200e-08 2
## SRSF1.2 1.016871e-12 0.6127839 0.143 0.321 3.721950e-08 2
## LYPLAL1 1.026942e-12 0.1366128 0.041 0.147 3.758815e-08 2
## POLR1C.1 1.030822e-12 0.1019980 0.033 0.134 3.773016e-08 2
## TRAFD1 1.038561e-12 0.3695883 0.056 0.172 3.801341e-08 2
## PPP1CB 1.043410e-12 0.6567613 0.156 0.346 3.819088e-08 2
## MRPL20-AS1 1.049828e-12 -0.9821326 0.012 0.095 3.842581e-08 2
## SFXN4.2 1.059742e-12 -0.4220547 0.020 0.109 3.878866e-08 2
## USP10 1.074366e-12 0.7332543 0.119 0.280 3.932394e-08 2
## LIN37.1 1.079316e-12 -0.4989218 0.012 0.095 3.950512e-08 2
## VPS33A 1.083419e-12 0.4073208 0.059 0.177 3.965528e-08 2
## EIF3J 1.096717e-12 0.5697644 0.101 0.248 4.014204e-08 2
## COA1 1.106815e-12 0.4432711 0.114 0.272 4.051164e-08 2
## GMCL1 1.109093e-12 0.4013662 0.075 0.205 4.059502e-08 2
## ZGPAT.1 1.120467e-12 0.6364931 0.078 0.211 4.101132e-08 2
## POLQ.2 1.135121e-12 -3.6958537 0.002 0.075 4.154771e-08 2
## PFKP.2 1.138874e-12 0.7052685 0.080 0.213 4.168508e-08 2
## TMEM167B 1.165006e-12 0.1717673 0.045 0.154 4.264155e-08 2
## SUPT20H.1 1.197594e-12 0.5737429 0.095 0.238 4.383433e-08 2
## CCDC97 1.230466e-12 0.1629669 0.045 0.154 4.503750e-08 2
## BOP1 1.252585e-12 0.4944630 0.041 0.146 4.584712e-08 2
## RPS6KA5.1 1.255587e-12 1.8371726 0.453 0.492 4.595699e-08 2
## CHKB 1.281939e-12 0.6863164 0.105 0.256 4.692152e-08 2
## RCSD1.1 1.293512e-12 0.4859154 0.292 0.592 4.734511e-08 2
## SRP54 1.295515e-12 0.7086646 0.113 0.270 4.741843e-08 2
## CCDC58.1 1.317500e-12 -1.1816501 0.012 0.095 4.822312e-08 2
## PDLIM1.1 1.318563e-12 -0.6267556 0.014 0.098 4.826202e-08 2
## WDR13 1.346884e-12 -0.1407505 0.027 0.122 4.929866e-08 2
## PHF6.1 1.364636e-12 0.2906043 0.066 0.188 4.994840e-08 2
## FNDC3B.2 1.369480e-12 2.4791956 0.290 0.224 5.012570e-08 2
## TM2D3 1.394417e-12 0.1626294 0.048 0.158 5.103844e-08 2
## CBFB.1 1.401729e-12 0.6642240 0.135 0.308 5.130609e-08 2
## CPSF3.1 1.433207e-12 0.4238870 0.041 0.146 5.245823e-08 2
## SLC39A6.1 1.463538e-12 -0.3932842 0.029 0.124 5.356841e-08 2
## LTV1 1.492954e-12 -0.2274959 0.042 0.148 5.464511e-08 2
## CPTP 1.505058e-12 -0.4067995 0.021 0.111 5.508813e-08 2
## ALG12 1.537485e-12 0.1487714 0.032 0.130 5.627503e-08 2
## CITED4.1 1.583337e-12 -0.5033041 0.021 0.111 5.795329e-08 2
## CCAR1 1.628199e-12 0.7613071 0.159 0.350 5.959536e-08 2
## TIGIT.2 1.630178e-12 -0.1842731 0.045 0.152 5.966776e-08 2
## TMX4.1 1.645434e-12 0.8023730 0.195 0.413 6.022619e-08 2
## DHX9.1 1.662839e-12 0.5847722 0.158 0.345 6.086323e-08 2
## LACTB2 1.667671e-12 0.2272925 0.023 0.114 6.104009e-08 2
## ALDOC 1.680916e-12 -2.7487130 0.003 0.077 6.152489e-08 2
## HAUS2.1 1.722384e-12 0.2412727 0.033 0.132 6.304271e-08 2
## C16orf87 1.725834e-12 -0.1909665 0.024 0.116 6.316897e-08 2
## NOM1 1.728754e-12 -0.4931443 0.020 0.108 6.327584e-08 2
## GTF3C5.1 1.827973e-12 0.7299882 0.045 0.153 6.690749e-08 2
## BLOC1S6 1.847642e-12 0.4740482 0.072 0.199 6.762740e-08 2
## RCE1 1.864938e-12 -0.2808403 0.023 0.113 6.826045e-08 2
## CLINT1 1.965909e-12 0.6076543 0.147 0.327 7.195620e-08 2
## CEACAM21.2 2.015470e-12 0.4474717 0.092 0.231 7.377022e-08 2
## KNL1.2 2.017937e-12 -0.9646127 0.018 0.104 7.386053e-08 2
## ZDHHC3.1 2.026883e-12 0.4669549 0.075 0.204 7.418795e-08 2
## MAPRE2.2 2.068888e-12 0.8120862 0.197 0.414 7.572542e-08 2
## TRAPPC2B 2.070680e-12 -0.4015283 0.018 0.105 7.579101e-08 2
## PPAN.1 2.073435e-12 0.9972330 0.081 0.214 7.589188e-08 2
## GMEB1 2.110523e-12 -0.3168926 0.023 0.113 7.724938e-08 2
## UBE2E3.1 2.140500e-12 0.2796731 0.056 0.170 7.834658e-08 2
## AP3D1 2.155619e-12 0.7196998 0.107 0.257 7.889997e-08 2
## BTBD9.1 2.174330e-12 2.4711501 0.337 0.293 7.958482e-08 2
## ZNF585A 2.191133e-12 -1.6464618 0.008 0.085 8.019986e-08 2
## KCTD18 2.212430e-12 -0.8660490 0.018 0.105 8.097936e-08 2
## PRPS2.2 2.212472e-12 0.1797367 0.033 0.132 8.098091e-08 2
## WDR36 2.246001e-12 0.2355787 0.050 0.160 8.220812e-08 2
## AKIRIN2.1 2.250379e-12 0.6197510 0.104 0.251 8.236837e-08 2
## SH3YL1 2.354125e-12 -1.3895485 0.014 0.096 8.616569e-08 2
## ORAI2 2.363825e-12 0.4313752 0.081 0.213 8.652071e-08 2
## TWISTNB.1 2.366378e-12 -0.2072697 0.035 0.134 8.661418e-08 2
## SOD2.1 2.394645e-12 0.6606289 0.119 0.278 8.764881e-08 2
## PRRC1 2.399955e-12 0.1402035 0.057 0.172 8.784315e-08 2
## GGH.2 2.407475e-12 -2.0306111 0.005 0.079 8.811839e-08 2
## SYVN1 2.423674e-12 -0.3784598 0.023 0.112 8.871132e-08 2
## CNOT8 2.446550e-12 0.5440040 0.104 0.252 8.954862e-08 2
## SLC35C2.1 2.456992e-12 0.4928556 0.069 0.193 8.993083e-08 2
## AP5M1 2.508839e-12 0.3854070 0.048 0.157 9.182852e-08 2
## LRRC47.1 2.551877e-12 0.2654667 0.053 0.164 9.340381e-08 2
## BIN3.1 2.576811e-12 0.2746880 0.044 0.149 9.431642e-08 2
## C6orf226.1 2.596251e-12 -0.3205195 0.017 0.102 9.502800e-08 2
## ARL3.1 2.642057e-12 0.8354011 0.044 0.149 9.670457e-08 2
## MIDN 2.681437e-12 0.3667694 0.095 0.234 9.814596e-08 2
## RUSC1.1 2.705886e-12 -0.2792024 0.021 0.110 9.904084e-08 2
## EIF2B3.1 2.731193e-12 0.5154394 0.059 0.175 9.996711e-08 2
## RIOK1 2.746782e-12 0.4456799 0.056 0.169 1.005377e-07 2
## MAGEH1 2.771558e-12 -0.7399849 0.015 0.099 1.014446e-07 2
## CTDSP2 2.782671e-12 0.2937450 0.066 0.187 1.018513e-07 2
## CEP55.2 2.811778e-12 -1.2193983 0.008 0.084 1.029167e-07 2
## CRIPT 2.865905e-12 0.1257775 0.050 0.159 1.048979e-07 2
## METTL17.1 2.883893e-12 0.7416197 0.077 0.205 1.055562e-07 2
## CHPT1.1 2.901708e-12 -0.1195873 0.029 0.123 1.062083e-07 2
## RPL39L.2 2.951007e-12 -3.6115468 0.002 0.072 1.080128e-07 2
## BORCS8 2.967218e-12 -0.3557891 0.021 0.109 1.086061e-07 2
## RDH11 2.967803e-12 0.4860986 0.056 0.169 1.086275e-07 2
## RMC1 2.991584e-12 0.4913620 0.048 0.157 1.094979e-07 2
## TOP1 3.011459e-12 0.7438976 0.202 0.425 1.102254e-07 2
## SEC63 3.014081e-12 0.6418970 0.132 0.299 1.103214e-07 2
## HSCB 3.042108e-12 -0.3303128 0.018 0.104 1.113472e-07 2
## HCCS.1 3.118201e-12 -0.1158965 0.018 0.104 1.141324e-07 2
## TYSND1 3.131935e-12 -0.5322821 0.017 0.101 1.146351e-07 2
## GSTM4 3.138305e-12 -0.3072513 0.014 0.095 1.148682e-07 2
## TCIRG1.1 3.185659e-12 0.8135987 0.158 0.344 1.166015e-07 2
## NAA80.1 3.190442e-12 -0.5591556 0.017 0.101 1.167766e-07 2
## UBE2D1.1 3.204254e-12 0.5785901 0.060 0.176 1.172821e-07 2
## ATAD2.2 3.238054e-12 0.4295860 0.078 0.205 1.185193e-07 2
## ORC3.1 3.337243e-12 0.2383582 0.042 0.146 1.221498e-07 2
## THEM6.1 3.337320e-12 -0.2713721 0.018 0.104 1.221526e-07 2
## IARS2 3.339153e-12 0.4192375 0.071 0.194 1.222197e-07 2
## FANCG.2 3.356362e-12 -0.5453068 0.009 0.087 1.228495e-07 2
## MSI2.1 3.388735e-12 2.3604937 0.337 0.295 1.240345e-07 2
## BET1 3.396043e-12 0.4226712 0.047 0.153 1.243020e-07 2
## PSAT1.2 3.411313e-12 -0.1827134 0.023 0.112 1.248609e-07 2
## TMEM33 3.441532e-12 0.1292784 0.050 0.158 1.259669e-07 2
## SMCO4.1 3.442671e-12 -0.2585848 0.026 0.117 1.260086e-07 2
## XPNPEP1.1 3.444172e-12 0.6256286 0.065 0.184 1.260636e-07 2
## ADORA2A 3.479124e-12 -0.5245366 0.021 0.109 1.273429e-07 2
## PML 3.497907e-12 0.5572234 0.081 0.212 1.280304e-07 2
## DALRD3 3.549946e-12 0.7764832 0.045 0.151 1.299351e-07 2
## CCDC82 3.556424e-12 0.3631364 0.063 0.181 1.301722e-07 2
## FNDC10 3.639004e-12 -0.7304535 0.009 0.087 1.331948e-07 2
## PCID2.1 3.706050e-12 0.6840153 0.072 0.197 1.356488e-07 2
## KIF11.2 3.870628e-12 -0.9669341 0.012 0.092 1.416727e-07 2
## NUP88.1 3.879934e-12 0.2923460 0.045 0.151 1.420133e-07 2
## IFNAR1 3.936022e-12 0.3827456 0.069 0.191 1.440663e-07 2
## MND1.2 3.953955e-12 -3.3908264 0.002 0.071 1.447227e-07 2
## WASHC1 3.959746e-12 1.0295166 0.113 0.266 1.449346e-07 2
## TMA16 3.997920e-12 -0.2882969 0.024 0.114 1.463319e-07 2
## NUP54.1 4.006965e-12 0.4601470 0.068 0.189 1.466629e-07 2
## XCL2.2 4.076106e-12 -1.3108162 0.015 0.097 1.491936e-07 2
## NUBP1 4.086034e-12 0.2589483 0.033 0.130 1.495570e-07 2
## NMT1 4.388097e-12 0.6048585 0.072 0.196 1.606131e-07 2
## MAP4K2 4.398577e-12 0.6530576 0.065 0.183 1.609967e-07 2
## EWSR1 4.405223e-12 0.4298591 0.305 0.612 1.612400e-07 2
## SPNS1.1 4.447129e-12 0.3782958 0.057 0.171 1.627738e-07 2
## RFNG 4.604434e-12 -0.1323813 0.023 0.111 1.685315e-07 2
## CENPS.2 4.643977e-12 -2.6565151 0.003 0.074 1.699789e-07 2
## MBD4 4.705594e-12 0.7241831 0.080 0.209 1.722341e-07 2
## LSR.1 4.803737e-12 -0.8008346 0.021 0.108 1.758264e-07 2
## PTS.1 4.836048e-12 0.5537652 0.054 0.165 1.770090e-07 2
## NFKBIZ.2 4.853881e-12 1.8456002 0.405 0.394 1.776617e-07 2
## KMT2C 4.860483e-12 2.0098656 0.402 0.406 1.779034e-07 2
## ZDHHC2 4.867471e-12 0.1685291 0.056 0.168 1.781592e-07 2
## ZNF749 4.889835e-12 -1.4060211 0.006 0.080 1.789777e-07 2
## WASHC5 4.911857e-12 0.5558850 0.065 0.184 1.797838e-07 2
## CARS.1 5.013916e-12 0.6667782 0.096 0.236 1.835194e-07 2
## TMEM115 5.030299e-12 -0.8152529 0.021 0.108 1.841190e-07 2
## ATXN7L3B 5.125878e-12 0.6220838 0.123 0.280 1.876174e-07 2
## EFTUD2 5.235221e-12 0.4409213 0.075 0.200 1.916196e-07 2
## PSTPIP1.1 5.290283e-12 0.9345475 0.144 0.318 1.936350e-07 2
## POC1A.2 5.302420e-12 -2.2226383 0.002 0.071 1.940792e-07 2
## TRA2B 5.325112e-12 0.5435625 0.244 0.496 1.949098e-07 2
## BIRC5.2 5.394482e-12 -2.2403080 0.003 0.074 1.974488e-07 2
## SIK3 5.397299e-12 1.9230688 0.411 0.417 1.975520e-07 2
## TNFSF8.2 5.517306e-12 0.4830109 0.232 0.463 2.019444e-07 2
## NELL2.1 5.628139e-12 0.3919499 0.104 0.244 2.060011e-07 2
## MRRF 5.662235e-12 0.4286061 0.044 0.147 2.072491e-07 2
## RIDA.1 5.705164e-12 -2.0746209 0.002 0.070 2.088204e-07 2
## ACOT2 5.743787e-12 -1.6717243 0.009 0.085 2.102341e-07 2
## ZNF598.1 5.857712e-12 0.1847201 0.030 0.123 2.144040e-07 2
## SIAH2 5.997889e-12 0.4966801 0.080 0.207 2.195347e-07 2
## TAZ 6.042696e-12 0.8696533 0.087 0.221 2.211748e-07 2
## ATG4D 6.080494e-12 -0.1964096 0.027 0.118 2.225582e-07 2
## DUS3L.1 6.132309e-12 0.6325744 0.041 0.142 2.244548e-07 2
## RAP1A.1 6.141051e-12 0.5366334 0.301 0.601 2.247747e-07 2
## REEP4.1 6.162382e-12 -0.3014288 0.021 0.107 2.255555e-07 2
## MRM3.1 6.309587e-12 -1.0172963 0.012 0.091 2.309435e-07 2
## PAM16 6.343588e-12 0.5415424 0.062 0.177 2.321880e-07 2
## CYLD.1 6.489095e-12 0.4522074 0.283 0.570 2.375139e-07 2
## ARHGAP30.2 6.508549e-12 0.5772132 0.253 0.512 2.382259e-07 2
## BAG5.2 6.525153e-12 0.2935365 0.050 0.157 2.388337e-07 2
## CARS2.2 6.591263e-12 0.7614536 0.071 0.192 2.412534e-07 2
## VAMP4.1 6.596493e-12 0.4132846 0.069 0.190 2.414448e-07 2
## THOC5 6.650781e-12 -0.8061589 0.023 0.110 2.434319e-07 2
## ATP8B4.2 6.706804e-12 2.6738732 0.259 0.190 2.454824e-07 2
## PRADC1.1 6.787429e-12 -0.6198924 0.008 0.082 2.484335e-07 2
## OGG1.1 6.853309e-12 0.1714576 0.026 0.115 2.508448e-07 2
## MBIP 6.899068e-12 -0.2316665 0.023 0.110 2.525197e-07 2
## PRPF4.1 7.003679e-12 0.1195658 0.026 0.115 2.563487e-07 2
## NUDT16.1 7.019421e-12 -2.0724252 0.008 0.082 2.569248e-07 2
## DCAF12 7.132097e-12 -0.1052184 0.033 0.128 2.610490e-07 2
## RIOX2 7.165040e-12 -0.1684348 0.035 0.131 2.622548e-07 2
## TRIM11.1 7.171004e-12 0.4381652 0.056 0.167 2.624731e-07 2
## KCNA3.2 7.371084e-12 0.4041405 0.066 0.184 2.697964e-07 2
## FO393401.1.1 7.399567e-12 -0.1912684 0.026 0.115 2.708389e-07 2
## CDCA5.2 7.424002e-12 -1.4406735 0.006 0.079 2.717333e-07 2
## TMEM42 7.448508e-12 -1.0593011 0.011 0.087 2.726303e-07 2
## CDK5RAP3 7.532906e-12 1.0371550 0.150 0.329 2.757194e-07 2
## VPS4B 7.681319e-12 0.6815203 0.114 0.266 2.811516e-07 2
## SQSTM1.1 7.691538e-12 1.2210090 0.292 0.588 2.815257e-07 2
## NT5DC1 7.718124e-12 0.8179854 0.117 0.272 2.824988e-07 2
## DCTN6.1 7.763434e-12 -1.4011682 0.008 0.082 2.841572e-07 2
## NUSAP1.2 7.782111e-12 -0.2973378 0.056 0.163 2.848408e-07 2
## PGS1 7.825965e-12 0.4346899 0.065 0.181 2.864460e-07 2
## OGFOD2 7.882229e-12 -0.4820318 0.017 0.099 2.885054e-07 2
## DDX21.1 7.886307e-12 0.6902557 0.173 0.368 2.886546e-07 2
## PCK2.1 7.992455e-12 -0.5088626 0.018 0.101 2.925399e-07 2
## BLCAP 8.057105e-12 0.5940389 0.062 0.176 2.949061e-07 2
## REPIN1 8.238634e-12 -0.5717792 0.026 0.114 3.015505e-07 2
## CENPV.1 8.333853e-12 -2.2179266 0.006 0.078 3.050357e-07 2
## TCF19.2 8.386811e-12 -0.3661285 0.017 0.098 3.069740e-07 2
## PLCB1.1 8.436701e-12 3.7369001 0.069 0.024 3.088001e-07 2
## SIKE1 8.476813e-12 0.3163116 0.053 0.161 3.102683e-07 2
## NRAS 8.592707e-12 0.2127765 0.048 0.153 3.145103e-07 2
## ZFAND3.1 8.765272e-12 2.1846974 0.355 0.325 3.208265e-07 2
## GPATCH11.1 8.774802e-12 -0.1130588 0.018 0.101 3.211753e-07 2
## SUN2.2 8.885329e-12 0.3834861 0.310 0.613 3.252208e-07 2
## PMM2 9.041813e-12 0.2897933 0.053 0.161 3.309484e-07 2
## FAM102A.1 9.144634e-12 0.6455942 0.141 0.312 3.347119e-07 2
## TAF1D 9.197970e-12 0.9275492 0.203 0.422 3.366641e-07 2
## DEF6.1 9.237981e-12 0.6831169 0.247 0.501 3.381286e-07 2
## PMS2.1 9.338415e-12 0.5824139 0.051 0.159 3.418047e-07 2
## ERCC5.1 9.426501e-12 0.5504342 0.138 0.304 3.450288e-07 2
## RCAN3AS 9.638983e-12 -1.0135763 0.014 0.092 3.528060e-07 2
## RRAGC 9.723585e-12 -0.3517169 0.030 0.122 3.559027e-07 2
## RAB5B.1 9.760262e-12 0.3845879 0.065 0.181 3.572451e-07 2
## FBXO45.1 9.893652e-12 -0.7527355 0.015 0.095 3.621275e-07 2
## RAD1.2 1.004651e-11 -0.4278852 0.018 0.100 3.677225e-07 2
## GBA 1.026861e-11 0.4839239 0.050 0.156 3.758517e-07 2
## TMEM99 1.035431e-11 -1.3357336 0.005 0.075 3.789885e-07 2
## AC011603.2.2 1.035933e-11 -0.5026524 0.015 0.095 3.791722e-07 2
## CSTF2.1 1.045495e-11 0.1676780 0.032 0.124 3.826722e-07 2
## P2RX4.1 1.067675e-11 0.6856327 0.080 0.206 3.907904e-07 2
## VPS16 1.070919e-11 0.3099940 0.048 0.152 3.919779e-07 2
## SLC39A8.2 1.086416e-11 0.8456104 0.111 0.260 3.976500e-07 2
## ZC3H13.1 1.087155e-11 0.8233354 0.146 0.320 3.979206e-07 2
## PTRH2.1 1.091076e-11 0.7028812 0.042 0.143 3.993556e-07 2
## CPSF7.1 1.106068e-11 0.2279524 0.059 0.170 4.048429e-07 2
## SLC66A2 1.129217e-11 0.6447642 0.053 0.160 4.133160e-07 2
## NDUFAF2 1.135108e-11 0.3092040 0.050 0.155 4.154723e-07 2
## IFT52 1.142448e-11 0.2590118 0.024 0.111 4.181590e-07 2
## TIA1 1.145969e-11 0.6110875 0.096 0.233 4.194474e-07 2
## AC006064.4.2 1.164053e-11 -0.7105839 0.006 0.078 4.260667e-07 2
## OBI1.1 1.174206e-11 -0.1281985 0.026 0.114 4.297828e-07 2
## TMEM102.1 1.178197e-11 -0.6550066 0.020 0.103 4.312436e-07 2
## PHF1.1 1.178907e-11 0.3271129 0.045 0.148 4.315034e-07 2
## HSPA5 1.186368e-11 0.3155867 0.359 0.696 4.342344e-07 2
## TBP 1.202569e-11 -0.3215761 0.029 0.119 4.401644e-07 2
## DCAF15.1 1.204326e-11 0.6909189 0.059 0.170 4.408075e-07 2
## TMEM273.2 1.206485e-11 0.4205436 0.102 0.241 4.415976e-07 2
## PDF.2 1.218484e-11 -1.1761655 0.011 0.086 4.459895e-07 2
## RANGAP1.2 1.228427e-11 0.3529493 0.038 0.134 4.496289e-07 2
## TSC22D3.2 1.239735e-11 0.6417461 0.215 0.436 4.537677e-07 2
## FAM89B 1.254876e-11 -1.8073981 0.006 0.077 4.593095e-07 2
## FNBP1 1.265326e-11 1.4186820 0.507 0.632 4.631346e-07 2
## STAT4.1 1.270330e-11 1.6359358 0.483 0.566 4.649663e-07 2
## ZNF444 1.276006e-11 0.1294943 0.032 0.124 4.670438e-07 2
## TRIM22.2 1.282761e-11 0.5399150 0.244 0.488 4.695160e-07 2
## IPO7.2 1.290709e-11 0.5634950 0.110 0.255 4.724252e-07 2
## STX17.1 1.311656e-11 0.3837661 0.054 0.162 4.800922e-07 2
## DHX36.1 1.322515e-11 0.7740186 0.153 0.332 4.840669e-07 2
## LINC00299.2 1.335197e-11 3.0802777 0.110 0.050 4.887087e-07 2
## NIF3L1.1 1.335468e-11 1.0112731 0.035 0.129 4.888081e-07 2
## CEP78.2 1.345406e-11 -0.3478603 0.023 0.108 4.924455e-07 2
## DNAJB12 1.352609e-11 0.4142724 0.054 0.162 4.950820e-07 2
## IRF2BPL.1 1.379897e-11 -0.7148301 0.015 0.094 5.050699e-07 2
## AP1B1.1 1.403309e-11 0.5166045 0.054 0.162 5.136392e-07 2
## CHMP1B 1.415063e-11 0.2126082 0.036 0.131 5.179413e-07 2
## MIER1.1 1.439450e-11 0.6287728 0.198 0.412 5.268673e-07 2
## LRCH4 1.451010e-11 0.6842571 0.078 0.202 5.310988e-07 2
## GFM1 1.459283e-11 0.3880420 0.057 0.167 5.341268e-07 2
## TBC1D4.1 1.474337e-11 0.6242253 0.066 0.182 5.396369e-07 2
## STX4.1 1.490411e-11 0.4612328 0.071 0.189 5.455201e-07 2
## SCCPDH.2 1.509880e-11 -0.3944045 0.012 0.088 5.526464e-07 2
## SPOCK2.1 1.523181e-11 0.5609692 0.283 0.564 5.575145e-07 2
## CAMK2G 1.530782e-11 0.7747310 0.093 0.228 5.602968e-07 2
## ETF1 1.546931e-11 0.5075120 0.107 0.249 5.662078e-07 2
## KIF15.2 1.561375e-11 -1.6909427 0.006 0.077 5.714943e-07 2
## KCTD20.1 1.562009e-11 0.4942344 0.071 0.189 5.717266e-07 2
## C11orf54 1.577534e-11 0.3229017 0.030 0.121 5.774090e-07 2
## ZNF688 1.585297e-11 0.1750203 0.029 0.118 5.802506e-07 2
## IFNAR2 1.590562e-11 0.7175996 0.182 0.380 5.821776e-07 2
## RBFA.2 1.598080e-11 0.3269113 0.027 0.115 5.849294e-07 2
## PUSL1.1 1.630232e-11 -0.8579003 0.011 0.085 5.966976e-07 2
## HAUS8.2 1.630855e-11 -0.5816954 0.009 0.082 5.969254e-07 2
## CAMK1.1 1.644467e-11 -0.6832064 0.017 0.096 6.019078e-07 2
## WDR5.2 1.646156e-11 -0.1205224 0.029 0.118 6.025260e-07 2
## TMEM120A 1.649063e-11 -0.3036450 0.011 0.085 6.035901e-07 2
## AKT2 1.651557e-11 0.3863705 0.048 0.151 6.045030e-07 2
## HPS6 1.673989e-11 -0.2516962 0.020 0.102 6.127134e-07 2
## XCL1.1 1.699150e-11 -0.7846891 0.033 0.123 6.219229e-07 2
## RNF34 1.729245e-11 0.6750255 0.089 0.219 6.329384e-07 2
## CRLF3.1 1.751091e-11 0.5922267 0.143 0.312 6.409345e-07 2
## FBXL3 1.755583e-11 0.1379060 0.042 0.141 6.425784e-07 2
## DDX24 1.765595e-11 0.6914626 0.254 0.516 6.462431e-07 2
## SYAP1 1.766168e-11 0.4980486 0.048 0.151 6.464527e-07 2
## CARD8 1.772337e-11 0.4943580 0.140 0.303 6.487109e-07 2
## SLC35C1 1.831389e-11 -1.0585455 0.014 0.091 6.703252e-07 2
## MAP1S 1.857967e-11 0.1227883 0.024 0.110 6.800529e-07 2
## CDKN3.2 1.861465e-11 -0.7525729 0.012 0.088 6.813334e-07 2
## DPH2 1.874907e-11 -1.2460790 0.011 0.085 6.862536e-07 2
## LEO1.1 1.907372e-11 0.5013745 0.066 0.182 6.981362e-07 2
## CLN3.1 1.944496e-11 0.7656238 0.080 0.204 7.117243e-07 2
## CFP.1 1.947183e-11 -0.8920212 0.014 0.091 7.127078e-07 2
## UBQLN4 1.974390e-11 -0.3194652 0.018 0.099 7.226660e-07 2
## ACY1 2.009632e-11 -0.1597029 0.014 0.090 7.355656e-07 2
## GUCD1.2 2.038725e-11 -0.2566909 0.023 0.107 7.462142e-07 2
## GOSR2 2.041038e-11 0.5711586 0.063 0.176 7.470606e-07 2
## POMGNT1 2.112714e-11 -1.1088380 0.012 0.087 7.732957e-07 2
## ADCK2 2.113471e-11 -0.8866388 0.009 0.082 7.735725e-07 2
## HBP1.1 2.139331e-11 0.4197281 0.093 0.225 7.830381e-07 2
## PANK2 2.145610e-11 0.5926419 0.086 0.213 7.853363e-07 2
## NKIRAS2 2.180973e-11 -0.1650585 0.024 0.109 7.982796e-07 2
## SASS6.2 2.186439e-11 -0.6136656 0.015 0.093 8.002804e-07 2
## CASP2 2.235741e-11 0.4250757 0.045 0.145 8.183258e-07 2
## TRAK2 2.235791e-11 0.5902259 0.090 0.219 8.183442e-07 2
## CEP63 2.248656e-11 0.5383329 0.056 0.163 8.230531e-07 2
## COX19 2.293366e-11 0.6974315 0.045 0.146 8.394177e-07 2
## VPS37A 2.304729e-11 0.3111414 0.038 0.132 8.435771e-07 2
## CDK2.2 2.339946e-11 -0.4534271 0.011 0.084 8.564671e-07 2
## SFT2D2 2.394944e-11 0.5736870 0.068 0.182 8.765973e-07 2
## FBXO5.2 2.410549e-11 -0.1222720 0.021 0.103 8.823091e-07 2
## PHYKPL.2 2.475503e-11 0.6506942 0.156 0.332 9.060835e-07 2
## BTG1.2 2.491677e-11 0.2077975 0.444 0.799 9.120035e-07 2
## FAF2 2.499360e-11 0.6992995 0.086 0.213 9.148156e-07 2
## PCCB 2.553986e-11 0.1048119 0.029 0.117 9.348099e-07 2
## COQ6 2.585728e-11 -0.5885802 0.017 0.095 9.464282e-07 2
## CHAF1A.2 2.597288e-11 0.5041860 0.038 0.132 9.506593e-07 2
## ARF4 2.606658e-11 0.6862791 0.218 0.442 9.540891e-07 2
## CCDC92 2.672186e-11 0.2142497 0.054 0.160 9.780737e-07 2
## SUCLA2 2.725261e-11 0.6654605 0.086 0.212 9.975002e-07 2
## PHLDA3.1 2.809128e-11 -0.2252998 0.032 0.121 1.028197e-06 2
## FUNDC1 2.812546e-11 -2.2529235 0.005 0.072 1.029448e-06 2
## GNPDA2 2.819994e-11 -1.3321028 0.014 0.089 1.032174e-06 2
## DPH1 2.833432e-11 -0.1280232 0.027 0.114 1.037093e-06 2
## WDR76.2 2.850486e-11 0.1245101 0.020 0.100 1.043335e-06 2
## ADSS.1 2.854260e-11 0.8413470 0.147 0.318 1.044716e-06 2
## TBC1D20 2.864756e-11 0.1574335 0.042 0.140 1.048558e-06 2
## AC079793.1.1 2.889792e-11 0.6350494 0.173 0.359 1.057722e-06 2
## TOPBP1.2 2.899283e-11 0.6253565 0.087 0.214 1.061196e-06 2
## SIRT7 2.905447e-11 0.5743356 0.077 0.198 1.063452e-06 2
## MGAT2 2.936629e-11 0.5498139 0.075 0.196 1.074865e-06 2
## GTF2F2 2.958565e-11 0.7260837 0.075 0.195 1.082894e-06 2
## FAM104B 3.000728e-11 -2.5362651 0.005 0.072 1.098327e-06 2
## EIF2B2 3.055117e-11 0.6068410 0.050 0.152 1.118234e-06 2
## CDCA8.2 3.056259e-11 -0.1843479 0.011 0.083 1.118652e-06 2
## DAPP1 3.105845e-11 0.4287411 0.044 0.142 1.136801e-06 2
## PXN.2 3.119039e-11 0.7723558 0.119 0.268 1.141631e-06 2
## MKNK2 3.171668e-11 0.9517755 0.189 0.395 1.160894e-06 2
## HNRNPU 3.235006e-11 0.3995712 0.358 0.707 1.184077e-06 2
## YTHDF1.1 3.291358e-11 0.2026308 0.045 0.144 1.204703e-06 2
## EML2 3.327211e-11 0.5650469 0.086 0.211 1.217826e-06 2
## ZFAND6.1 3.328269e-11 0.7413402 0.147 0.316 1.218213e-06 2
## KLRD1.2 3.336221e-11 -0.1773250 0.054 0.157 1.221124e-06 2
## GBP3 3.350163e-11 0.2856188 0.054 0.159 1.226227e-06 2
## L3MBTL2.1 3.410248e-11 0.6112869 0.035 0.126 1.248219e-06 2
## PPIL4 3.429208e-11 0.7047991 0.101 0.237 1.255159e-06 2
## SIPA1.1 3.430109e-11 0.7711799 0.123 0.275 1.255489e-06 2
## FOXM1.2 3.521868e-11 -2.1861679 0.003 0.069 1.289074e-06 2
## NFKBIL1 3.554811e-11 -0.1162834 0.021 0.102 1.301132e-06 2
## RAB6A 3.593320e-11 0.6750954 0.164 0.346 1.315227e-06 2
## REEP3.2 3.700415e-11 0.7223148 0.087 0.214 1.354426e-06 2
## RTL8A 3.722626e-11 -0.7019286 0.020 0.100 1.362555e-06 2
## AURKB.2 3.734145e-11 -1.9764129 0.005 0.071 1.366772e-06 2
## TMEM209.1 3.741882e-11 0.2062873 0.048 0.149 1.369604e-06 2
## TMEM69.1 3.766641e-11 0.1188883 0.021 0.102 1.378666e-06 2
## EHMT2.1 3.784910e-11 0.4194032 0.060 0.169 1.385353e-06 2
## BROX 3.787178e-11 0.3610794 0.066 0.179 1.386183e-06 2
## MARS 3.808841e-11 0.9649148 0.098 0.233 1.394112e-06 2
## RO60 3.892412e-11 0.5735461 0.083 0.206 1.424701e-06 2
## RAD51AP1.2 3.896321e-11 -0.6730315 0.014 0.088 1.426131e-06 2
## STAU1 3.919345e-11 0.6464912 0.132 0.290 1.434559e-06 2
## ARHGAP1 3.920397e-11 0.6812805 0.066 0.179 1.434944e-06 2
## PDCD6IP 3.938169e-11 0.7241434 0.137 0.299 1.441449e-06 2
## CCDC137.1 3.997884e-11 0.5690645 0.051 0.154 1.463305e-06 2
## LIPA 4.036081e-11 0.6842350 0.045 0.144 1.477286e-06 2
## NDUFAF1.1 4.117713e-11 -0.2372004 0.015 0.091 1.507165e-06 2
## RITA1.1 4.159804e-11 -0.2163263 0.014 0.088 1.522571e-06 2
## RNF10 4.199428e-11 0.8472652 0.129 0.286 1.537075e-06 2
## PEA15.1 4.305940e-11 -1.0004837 0.008 0.077 1.576060e-06 2
## RAVER1.1 4.324738e-11 0.3897485 0.045 0.144 1.582941e-06 2
## SART3 4.329566e-11 0.3963025 0.057 0.164 1.584708e-06 2
## TAMM41 4.331919e-11 0.2599022 0.036 0.128 1.585569e-06 2
## ALKBH6 4.397862e-11 0.3128722 0.047 0.145 1.609705e-06 2
## AMDHD2 4.412743e-11 0.4278685 0.029 0.115 1.615152e-06 2
## PCGF6.2 4.461325e-11 -0.2539123 0.021 0.102 1.632934e-06 2
## PAK1IP1.1 4.477759e-11 0.1617102 0.030 0.118 1.638949e-06 2
## HDHD2 4.506678e-11 0.1708893 0.045 0.143 1.649534e-06 2
## MCM10.2 4.547325e-11 -0.7396781 0.006 0.074 1.664412e-06 2
## C21orf91.2 4.635491e-11 0.7753324 0.101 0.236 1.696682e-06 2
## SNAI3 4.657153e-11 -0.4929577 0.021 0.101 1.704611e-06 2
## LINC002481.2 4.690305e-11 -0.4156668 0.012 0.085 1.716745e-06 2
## TRMT1 4.786182e-11 0.8170366 0.059 0.166 1.751838e-06 2
## PAG1.1 4.787963e-11 1.9650252 0.424 0.454 1.752490e-06 2
## BTN3A3 4.799171e-11 0.1937617 0.051 0.153 1.756593e-06 2
## DNAJB6 4.812518e-11 0.7957293 0.110 0.251 1.761478e-06 2
## UBALD1 4.815713e-11 -0.2254048 0.020 0.099 1.762647e-06 2
## CFDP1 4.819735e-11 0.7433751 0.114 0.259 1.764119e-06 2
## CIP2A.2 5.002919e-11 -0.2480842 0.012 0.085 1.831168e-06 2
## COPS7A 5.026472e-11 0.1733688 0.038 0.130 1.839789e-06 2
## PSMD5 5.091201e-11 0.1813917 0.048 0.148 1.863481e-06 2
## PCGF5.1 5.164489e-11 0.6729636 0.161 0.338 1.890306e-06 2
## CRYZ 5.185076e-11 -0.8466076 0.012 0.085 1.897841e-06 2
## P2RY10 5.246470e-11 0.2256003 0.056 0.160 1.920313e-06 2
## NAPG 5.247119e-11 0.3724626 0.056 0.160 1.920551e-06 2
## GTF2E2 5.254525e-11 0.5458900 0.087 0.213 1.923261e-06 2
## AC026347.1.1 5.337329e-11 0.2166516 0.041 0.135 1.953569e-06 2
## SH3GL1 5.421795e-11 0.7059837 0.069 0.183 1.984485e-06 2
## RACGAP1.2 5.425558e-11 -0.4598056 0.024 0.106 1.985863e-06 2
## HYOU1 5.517791e-11 0.4562359 0.077 0.194 2.019622e-06 2
## C2orf49 5.522552e-11 -1.0062334 0.012 0.085 2.021364e-06 2
## GNL1.1 5.535903e-11 0.5605390 0.074 0.190 2.026251e-06 2
## PELO 5.543674e-11 -0.1099583 0.027 0.112 2.029095e-06 2
## UBE2Q2 5.550345e-11 0.6343171 0.089 0.215 2.031537e-06 2
## DUSP2.2 5.585256e-11 0.4426761 0.110 0.247 2.044316e-06 2
## MTFMT 5.761038e-11 -0.1439848 0.021 0.101 2.108655e-06 2
## TEDC1.2 5.808440e-11 -1.2088414 0.009 0.079 2.126005e-06 2
## MFSD12.1 5.841341e-11 0.4279375 0.053 0.155 2.138048e-06 2
## RNF8.1 5.874079e-11 0.6198793 0.042 0.138 2.150030e-06 2
## NEDD1.2 5.899705e-11 0.2739302 0.050 0.150 2.159410e-06 2
## KLF3.2 5.980912e-11 0.1965988 0.038 0.130 2.189134e-06 2
## PRR14 5.989216e-11 0.3491761 0.038 0.130 2.192173e-06 2
## GPALPP1 6.006701e-11 0.1171516 0.033 0.122 2.198573e-06 2
## CARD8-AS1 6.069408e-11 -0.1258308 0.020 0.098 2.221525e-06 2
## ZCCHC8 6.117264e-11 0.7951924 0.080 0.200 2.239041e-06 2
## TRMT61B 6.145793e-11 -0.1432292 0.023 0.103 2.249483e-06 2
## WAC-AS1.1 6.160558e-11 0.3593757 0.056 0.160 2.254888e-06 2
## HK1 6.196347e-11 0.7388959 0.149 0.318 2.267987e-06 2
## IDS.1 6.262482e-11 0.6151478 0.233 0.464 2.292194e-06 2
## TMEM251 6.323517e-11 -1.8728593 0.003 0.067 2.314534e-06 2
## USP30-AS1 6.339167e-11 -1.9273361 0.006 0.073 2.320262e-06 2
## PPP4R2 6.409316e-11 0.7777799 0.140 0.302 2.345938e-06 2
## ATP13A1 6.516354e-11 0.6191101 0.053 0.154 2.385116e-06 2
## AKT1S1 6.721369e-11 -2.5866378 0.003 0.067 2.460155e-06 2
## TP53I13 6.731947e-11 0.8102876 0.071 0.185 2.464027e-06 2
## MCMBP.1 6.738995e-11 0.7072184 0.062 0.169 2.466607e-06 2
## ZNF787 6.767769e-11 0.1337675 0.030 0.116 2.477139e-06 2
## ZDHHC16.1 6.818926e-11 0.1129611 0.020 0.098 2.495863e-06 2
## PXMP4 6.935155e-11 -0.7340607 0.014 0.087 2.538405e-06 2
## CHCHD4 6.957777e-11 -0.5911618 0.011 0.081 2.546686e-06 2
## DHRS1.1 7.146081e-11 -0.1542808 0.027 0.111 2.615609e-06 2
## GNB1 7.545792e-11 0.6090669 0.253 0.508 2.761911e-06 2
## PDCD4.1 7.739770e-11 0.4017108 0.289 0.563 2.832911e-06 2
## RELB.1 8.009648e-11 0.6339888 0.081 0.201 2.931691e-06 2
## TSPAN17.1 8.027821e-11 -0.5099954 0.020 0.097 2.938343e-06 2
## JMJD7.1 8.045530e-11 -0.2156869 0.026 0.108 2.944825e-06 2
## BYSL 8.059721e-11 -0.6697228 0.014 0.087 2.950019e-06 2
## MTERF4 8.160218e-11 0.3823402 0.063 0.172 2.986803e-06 2
## UBAP2L.1 8.228117e-11 0.6551788 0.096 0.227 3.011655e-06 2
## HSPA1B 8.247845e-11 0.5837451 0.050 0.149 3.018876e-06 2
## B3GNT2.1 8.482703e-11 0.4399176 0.068 0.179 3.104839e-06 2
## OTUD5 8.684326e-11 0.4346135 0.083 0.203 3.178637e-06 2
## ZNF277.1 8.753644e-11 0.5797996 0.081 0.200 3.204009e-06 2
## ATAD5.2 8.862691e-11 0.4198158 0.039 0.131 3.243922e-06 2
## PRKAR1B.1 8.922545e-11 0.6265793 0.071 0.184 3.265830e-06 2
## RNF25 9.027957e-11 -0.4712509 0.020 0.097 3.304413e-06 2
## NAALADL1 9.149211e-11 -0.6114323 0.014 0.086 3.348794e-06 2
## CYTH2 9.187025e-11 0.2232453 0.044 0.138 3.362635e-06 2
## ZFPL1.1 9.359820e-11 -0.1428225 0.030 0.115 3.425881e-06 2
## ZDHHC18 9.557586e-11 0.2187817 0.038 0.128 3.498268e-06 2
## CBLB.1 9.660882e-11 1.8050217 0.459 0.529 3.536076e-06 2
## TRIM38.1 9.704918e-11 0.7014707 0.116 0.257 3.552194e-06 2
## INTS14 9.735124e-11 0.1247317 0.026 0.108 3.563250e-06 2
## EXOC4.1 9.764918e-11 1.9452633 0.409 0.433 3.574155e-06 2
## AC079781.5 9.828273e-11 0.8292818 0.101 0.234 3.597345e-06 2
## C12orf49 9.877420e-11 0.2654935 0.032 0.118 3.615333e-06 2
## CWC27 9.912400e-11 0.8556383 0.090 0.216 3.628137e-06 2
## RAB39B 1.011062e-10 -0.7981828 0.009 0.078 3.700689e-06 2
## CCDC43 1.031019e-10 0.1066967 0.032 0.118 3.773737e-06 2
## GNB5 1.031921e-10 0.4658190 0.056 0.158 3.777038e-06 2
## DNAJC17.1 1.036799e-10 0.4329557 0.042 0.136 3.794891e-06 2
## TATDN3 1.041914e-10 0.1971064 0.026 0.107 3.813614e-06 2
## EIF4G2.1 1.052904e-10 0.2977301 0.350 0.671 3.853841e-06 2
## RAPGEF1.1 1.071824e-10 2.0580265 0.358 0.339 3.923088e-06 2
## DCP2 1.076893e-10 0.7478672 0.113 0.253 3.941644e-06 2
## MCCC2 1.102315e-10 0.3790555 0.054 0.156 4.034695e-06 2
## CDCA2.2 1.106973e-10 -1.3577743 0.008 0.074 4.051743e-06 2
## CYB5R1 1.113314e-10 -0.3767711 0.021 0.099 4.074952e-06 2
## LINC00909 1.114397e-10 -0.5785615 0.015 0.088 4.078914e-06 2
## KRT7.1 1.120581e-10 -2.0577087 0.006 0.071 4.101551e-06 2
## MTG1 1.145771e-10 0.5803432 0.081 0.200 4.193750e-06 2
## UBXN11.2 1.158627e-10 0.7778073 0.080 0.197 4.240808e-06 2
## AKAP13.1 1.170431e-10 1.5911740 0.483 0.588 4.284011e-06 2
## ANAPC7 1.176745e-10 0.5958273 0.042 0.136 4.307123e-06 2
## ERCC3 1.179318e-10 0.4606343 0.063 0.170 4.316538e-06 2
## TMEM97.2 1.181612e-10 -3.9272330 0.002 0.062 4.324937e-06 2
## PARPBP.2 1.199216e-10 -0.1542117 0.017 0.091 4.389369e-06 2
## BRI3BP.2 1.200642e-10 0.1272145 0.038 0.128 4.394592e-06 2
## UPF3B 1.202834e-10 0.7223326 0.051 0.151 4.402612e-06 2
## EML3 1.224756e-10 0.3737185 0.056 0.158 4.482851e-06 2
## RABL3 1.237717e-10 -0.1623459 0.029 0.112 4.530292e-06 2
## FAM114A2 1.238023e-10 -0.1122606 0.024 0.104 4.531412e-06 2
## ZNF544.1 1.253537e-10 -0.3442658 0.021 0.099 4.588195e-06 2
## SCAF1.1 1.255054e-10 0.3448995 0.032 0.117 4.593749e-06 2
## IMMP1L.1 1.271846e-10 0.3835511 0.045 0.140 4.655212e-06 2
## TMEM254 1.274328e-10 -0.6173469 0.011 0.080 4.664296e-06 2
## MARCH5 1.274537e-10 0.2726794 0.059 0.162 4.665060e-06 2
## GOT1.1 1.296079e-10 0.9459826 0.038 0.128 4.743909e-06 2
## SEPTIN9.1 1.305972e-10 0.3314504 0.394 0.749 4.780120e-06 2
## EIF2AK2.1 1.307787e-10 0.7025682 0.078 0.195 4.786763e-06 2
## ARSA.1 1.369713e-10 0.5532917 0.054 0.155 5.013422e-06 2
## HEBP1.2 1.376520e-10 -1.8778354 0.002 0.062 5.038337e-06 2
## NCBP3.1 1.387648e-10 0.6456459 0.069 0.180 5.079070e-06 2
## EVA1B.1 1.396407e-10 -0.2005752 0.024 0.104 5.111129e-06 2
## LINC01578 1.403417e-10 0.5076107 0.310 0.611 5.136785e-06 2
## CPB2-AS1.1 1.408877e-10 0.3462178 0.051 0.150 5.156771e-06 2
## INTS5 1.429499e-10 -0.7953169 0.012 0.082 5.232253e-06 2
## SLC41A3 1.449207e-10 -0.2796894 0.023 0.101 5.304388e-06 2
## POLD1.2 1.490310e-10 -0.2508146 0.024 0.104 5.454833e-06 2
## ATAD1 1.491544e-10 0.6382036 0.093 0.219 5.459350e-06 2
## MBD2.1 1.528987e-10 0.5314007 0.095 0.221 5.596398e-06 2
## SLA2.1 1.535028e-10 0.7409928 0.098 0.228 5.618510e-06 2
## TIMM8A 1.579818e-10 -2.1522664 0.005 0.068 5.782449e-06 2
## DNAL4 1.581025e-10 -2.1604826 0.005 0.068 5.786868e-06 2
## NUP93.1 1.599315e-10 0.4856752 0.060 0.164 5.853811e-06 2
## CTNNBIP1.1 1.603587e-10 0.5544414 0.033 0.119 5.869449e-06 2
## NADK 1.617715e-10 0.2647797 0.041 0.132 5.921160e-06 2
## SRSF2.1 1.663298e-10 0.5584382 0.322 0.617 6.088002e-06 2
## ZNF414 1.680097e-10 0.6196157 0.053 0.152 6.149492e-06 2
## TCP11L2.2 1.698673e-10 0.2264650 0.050 0.147 6.217482e-06 2
## APOL2.1 1.738605e-10 0.3670601 0.044 0.137 6.363643e-06 2
## RXRB 1.740565e-10 0.1274983 0.035 0.122 6.370818e-06 2
## LINC00339 1.753589e-10 0.1369608 0.026 0.106 6.418485e-06 2
## XRCC1.1 1.759268e-10 0.5068978 0.044 0.136 6.439271e-06 2
## ITPRIPL1.1 1.760072e-10 0.4305635 0.020 0.095 6.442214e-06 2
## KIAA0825.1 1.772499e-10 2.6564332 0.209 0.142 6.487702e-06 2
## EDEM2 1.789741e-10 0.5458102 0.066 0.174 6.550811e-06 2
## RMI2.2 1.798124e-10 0.8904543 0.066 0.174 6.581494e-06 2
## DUSP1.2 1.800574e-10 0.3105242 0.373 0.668 6.590462e-06 2
## OIP5.2 1.826938e-10 -1.7479664 0.005 0.067 6.686958e-06 2
## SMC6.1 1.854215e-10 0.7555793 0.116 0.256 6.786799e-06 2
## MPV17L2 1.856270e-10 -0.4362155 0.014 0.084 6.794320e-06 2
## ATP6V0A2 1.878694e-10 0.5233733 0.047 0.142 6.876396e-06 2
## BIRC2 1.881739e-10 0.4472146 0.060 0.164 6.887542e-06 2
## IFT57 1.884405e-10 0.1132687 0.044 0.136 6.897297e-06 2
## TMED7 1.888601e-10 0.2928360 0.047 0.141 6.912657e-06 2
## ING3 1.918381e-10 0.1212984 0.026 0.106 7.021657e-06 2
## KIN 1.924757e-10 0.6662909 0.050 0.147 7.044996e-06 2
## WBP4 1.942634e-10 0.2377942 0.026 0.106 7.110429e-06 2
## WEE1.2 1.949599e-10 0.2502147 0.030 0.113 7.135923e-06 2
## STX8.2 1.985353e-10 0.6527139 0.168 0.345 7.266789e-06 2
## MIS18A.2 2.029122e-10 -0.4765604 0.009 0.076 7.426994e-06 2
## SLC29A1.2 2.051824e-10 -1.2208097 0.006 0.070 7.510086e-06 2
## TFAP4.2 2.082846e-10 -1.1896590 0.011 0.079 7.623634e-06 2
## STYX 2.083957e-10 0.5227080 0.066 0.173 7.627699e-06 2
## GCAT.1 2.118985e-10 -0.4169357 0.005 0.067 7.755909e-06 2
## KMT5A.2 2.130087e-10 0.4175982 0.035 0.121 7.796546e-06 2
## ME2.1 2.139765e-10 0.8023187 0.114 0.254 7.831967e-06 2
## TTC4 2.143652e-10 -0.5196696 0.014 0.084 7.846194e-06 2
## WAPL 2.159166e-10 0.7848988 0.152 0.318 7.902979e-06 2
## RNF138.1 2.173856e-10 0.8125160 0.096 0.223 7.956748e-06 2
## IL10RB 2.177388e-10 0.4964199 0.063 0.169 7.969676e-06 2
## PDLIM7.1 2.193270e-10 -1.4523015 0.009 0.075 8.027807e-06 2
## UBP1 2.212683e-10 0.4026525 0.050 0.146 8.098862e-06 2
## CETN3.1 2.256610e-10 -0.4394605 0.011 0.078 8.259645e-06 2
## SLC30A5 2.260491e-10 0.6262161 0.057 0.159 8.273849e-06 2
## MRPS9 2.272099e-10 0.4232625 0.054 0.153 8.316337e-06 2
## ARID5A 2.284383e-10 0.2601519 0.050 0.146 8.361300e-06 2
## TMEM187 2.297130e-10 -2.6251392 0.003 0.064 8.407956e-06 2
## C18orf21 2.313290e-10 -0.1424090 0.023 0.100 8.467103e-06 2
## FUCA2 2.355994e-10 -0.4894823 0.011 0.078 8.623408e-06 2
## LY96.1 2.362259e-10 -1.4441347 0.006 0.070 8.646342e-06 2
## PLOD3 2.382305e-10 0.3956266 0.038 0.126 8.719713e-06 2
## CCNB1.2 2.387203e-10 -3.4244596 0.003 0.063 8.737642e-06 2
## TTC28-AS1 2.395289e-10 0.2858917 0.030 0.113 8.767236e-06 2
## HEMK1 2.402973e-10 0.5707859 0.045 0.138 8.795361e-06 2
## MANEA.2 2.417266e-10 0.1778999 0.033 0.118 8.847677e-06 2
## PUS7L 2.419562e-10 0.2755581 0.045 0.138 8.856079e-06 2
## RARS 2.423136e-10 0.3391527 0.048 0.143 8.869163e-06 2
## EGLN3.1 2.444528e-10 0.3907476 0.098 0.219 8.947462e-06 2
## TTC37 2.460852e-10 0.6056043 0.081 0.198 9.007211e-06 2
## ACBD3 2.569679e-10 0.7223309 0.068 0.175 9.405538e-06 2
## IRAK4.1 2.581692e-10 0.8279456 0.101 0.230 9.449511e-06 2
## SLC22A18.1 2.583156e-10 0.2497710 0.027 0.107 9.454866e-06 2
## DGKA.2 2.585744e-10 0.7223397 0.211 0.419 9.464340e-06 2
## PWWP2A 2.589302e-10 0.4761448 0.057 0.157 9.477363e-06 2
## INTS8 2.665150e-10 0.7009844 0.059 0.160 9.754984e-06 2
## ZNF720 2.667802e-10 0.5875515 0.056 0.155 9.764689e-06 2
## BRCA2.2 2.691227e-10 0.1423700 0.029 0.110 9.850429e-06 2
## SNX18.1 2.697530e-10 -2.0499090 0.008 0.072 9.873499e-06 2
## NLE1 2.749912e-10 -0.2401132 0.014 0.083 1.006523e-05 2
## ZRANB2 2.750810e-10 0.8709029 0.185 0.375 1.006852e-05 2
## DNAJC30 2.768697e-10 -0.6276909 0.014 0.083 1.013398e-05 2
## BRIP1.2 2.775989e-10 -0.1249872 0.027 0.107 1.016067e-05 2
## ELAC2.1 2.853257e-10 0.7184747 0.057 0.157 1.044349e-05 2
## MAT2A 2.872272e-10 0.8236239 0.161 0.331 1.051309e-05 2
## PNO1 2.889793e-10 0.2362510 0.030 0.112 1.057722e-05 2
## ABRAXAS1 2.904019e-10 -0.3378339 0.023 0.099 1.062929e-05 2
## CSNK2A2 2.914357e-10 0.6452669 0.069 0.177 1.066713e-05 2
## HEXD 2.962586e-10 0.4796450 0.060 0.162 1.084366e-05 2
## CDC42EP3.2 2.967796e-10 0.5593195 0.086 0.204 1.086273e-05 2
## DYNC1LI1 3.080401e-10 0.5100998 0.060 0.162 1.127488e-05 2
## AZI2 3.118557e-10 0.2785892 0.053 0.149 1.141454e-05 2
## PRXL2C 3.132756e-10 -1.3454732 0.008 0.071 1.146651e-05 2
## NPAT.1 3.134532e-10 0.8469200 0.090 0.211 1.147301e-05 2
## MELK.2 3.146932e-10 -0.3762192 0.017 0.088 1.151840e-05 2
## INO80C 3.152564e-10 -0.6917510 0.006 0.069 1.153902e-05 2
## STAP1.1 3.158638e-10 -0.1539196 0.023 0.099 1.156125e-05 2
## YY1AP1 3.167995e-10 0.7346188 0.089 0.209 1.159549e-05 2
## CCDC18-AS1 3.192216e-10 0.7704056 0.113 0.249 1.168415e-05 2
## LMBRD1 3.200590e-10 0.7010522 0.141 0.297 1.171480e-05 2
## SNHG30 3.209611e-10 0.1047487 0.017 0.088 1.174782e-05 2
## SPC25.2 3.249937e-10 -1.7467547 0.005 0.066 1.189542e-05 2
## BLOC1S5 3.266342e-10 0.3540080 0.027 0.107 1.195547e-05 2
## ACSL3 3.294958e-10 0.3422309 0.060 0.162 1.206021e-05 2
## TBC1D10A 3.316708e-10 0.8223673 0.084 0.202 1.213982e-05 2
## MED24.1 3.330257e-10 0.2641211 0.038 0.124 1.218941e-05 2
## SQLE.1 3.344914e-10 -0.3248847 0.015 0.085 1.224305e-05 2
## MVK.1 3.461147e-10 -0.5545926 0.008 0.071 1.266849e-05 2
## EIF1AD 3.470318e-10 -0.2062316 0.015 0.085 1.270206e-05 2
## UBE2E1.2 3.484779e-10 0.8490974 0.078 0.192 1.275499e-05 2
## SPATS2L.1 3.581141e-10 0.3080698 0.044 0.134 1.310769e-05 2
## CCDC61 3.583857e-10 -0.1771202 0.020 0.093 1.311763e-05 2
## CDKN2AIP.1 3.598950e-10 0.5526855 0.059 0.160 1.317288e-05 2
## GET1.1 3.650705e-10 -0.1607272 0.021 0.096 1.336231e-05 2
## PAPSS1.1 3.671248e-10 0.3599309 0.075 0.186 1.343750e-05 2
## ASCC2 3.684866e-10 0.7559417 0.071 0.180 1.348735e-05 2
## KDM1A.1 3.695484e-10 0.5388539 0.075 0.187 1.352621e-05 2
## ATP6V1H 3.706067e-10 0.8228725 0.110 0.245 1.356495e-05 2
## NMD3 3.742558e-10 0.2132966 0.044 0.135 1.369851e-05 2
## HMGB3.2 3.750163e-10 -0.4228295 0.012 0.079 1.372635e-05 2
## MTAP.1 3.770114e-10 0.8884199 0.147 0.307 1.379937e-05 2
## ICOS.1 3.788778e-10 0.7332662 0.135 0.285 1.386769e-05 2
## BRD4.1 3.812760e-10 0.9194488 0.173 0.351 1.395546e-05 2
## MPPE1 3.875141e-10 0.3247855 0.051 0.147 1.418379e-05 2
## EMC2.1 3.904343e-10 0.4411520 0.039 0.127 1.429068e-05 2
## TCTN3 3.911514e-10 0.7965315 0.048 0.142 1.431692e-05 2
## IRAK1.1 3.935070e-10 0.3397032 0.047 0.139 1.440314e-05 2
## C1orf52 3.947060e-10 0.7707200 0.087 0.207 1.444703e-05 2
## MAPKAPK5 3.978077e-10 0.4680447 0.051 0.147 1.456056e-05 2
## PEX19 3.999382e-10 0.1389282 0.035 0.119 1.463854e-05 2
## DSN1.2 4.137192e-10 0.6894082 0.026 0.104 1.514295e-05 2
## CTNNBL1 4.192814e-10 0.6443959 0.078 0.192 1.534654e-05 2
## RANBP2.1 4.221354e-10 0.8009741 0.165 0.338 1.545100e-05 2
## MT1E.1 4.234523e-10 0.2315253 0.021 0.096 1.549920e-05 2
## AP1G2.1 4.264187e-10 0.8926970 0.143 0.300 1.560778e-05 2
## HAGHL.1 4.307456e-10 -0.1431324 0.014 0.082 1.576615e-05 2
## NUP58.2 4.372891e-10 0.7063685 0.113 0.247 1.600566e-05 2
## TMEM9.1 4.446969e-10 -0.3174306 0.012 0.079 1.627680e-05 2
## MSL3 4.454226e-10 0.6112591 0.062 0.163 1.630336e-05 2
## CHORDC1.1 4.459891e-10 0.6433567 0.096 0.221 1.632409e-05 2
## TMEM8A 4.488327e-10 0.5657330 0.045 0.136 1.642818e-05 2
## TARDBP 4.579079e-10 0.5155366 0.069 0.176 1.676034e-05 2
## FBH1 4.586404e-10 0.3403163 0.048 0.141 1.678715e-05 2
## MED1 4.645042e-10 0.5964037 0.059 0.159 1.700178e-05 2
## MTIF2 4.659306e-10 -0.1684637 0.021 0.095 1.705399e-05 2
## ZNF800 4.695377e-10 0.8457268 0.147 0.306 1.718602e-05 2
## NDUFV3 4.722121e-10 0.6505061 0.060 0.161 1.728391e-05 2
## C19orf12.1 4.774062e-10 0.8958139 0.092 0.212 1.747402e-05 2
## SLC25A24 4.780846e-10 0.5993248 0.066 0.171 1.749885e-05 2
## DIPK1A.1 4.812466e-10 0.6383717 0.093 0.215 1.761459e-05 2
## MAGT1.1 4.856838e-10 0.6848331 0.087 0.205 1.777700e-05 2
## MECR.1 4.905401e-10 -0.6444709 0.008 0.071 1.795475e-05 2
## MFGE8.1 4.996544e-10 -2.4729368 0.003 0.062 1.828835e-05 2
## NUF2.2 5.042160e-10 -0.9076631 0.012 0.079 1.845532e-05 2
## FBRS.1 5.051974e-10 0.7364172 0.099 0.225 1.849124e-05 2
## ZBTB24 5.078354e-10 0.5196091 0.062 0.163 1.858779e-05 2
## PTPN11 5.080860e-10 0.8255784 0.135 0.286 1.859697e-05 2
## ALDH6A1 5.089180e-10 -1.5340157 0.008 0.070 1.862742e-05 2
## AEN 5.090380e-10 0.1118344 0.030 0.111 1.863181e-05 2
## KLRC4.2 5.091510e-10 0.1598336 0.075 0.183 1.863595e-05 2
## DLAT 5.254674e-10 -0.1312250 0.023 0.097 1.923316e-05 2
## N4BP2L1 5.267855e-10 0.2674009 0.056 0.153 1.928140e-05 2
## ZNF76 5.345526e-10 -0.1487560 0.021 0.095 1.956570e-05 2
## DYNC1I2.1 5.348678e-10 0.4382889 0.056 0.153 1.957723e-05 2
## FAM210A 5.352378e-10 0.5817951 0.032 0.113 1.959077e-05 2
## ALAD 5.372353e-10 0.2257105 0.026 0.103 1.966389e-05 2
## ALKBH2.1 5.420955e-10 -2.2385513 0.003 0.061 1.984178e-05 2
## AP5S1 5.444483e-10 -1.9227996 0.006 0.067 1.992790e-05 2
## TRIOBP.1 5.503498e-10 0.6802350 0.041 0.128 2.014390e-05 2
## PREPL 5.507286e-10 0.5845272 0.059 0.158 2.015777e-05 2
## CASTOR1 5.514090e-10 -1.0772408 0.011 0.076 2.018267e-05 2
## RABEP1 5.603827e-10 0.6949041 0.113 0.246 2.051113e-05 2
## TBL3 5.605405e-10 0.5229145 0.038 0.123 2.051690e-05 2
## ZNF384 5.619768e-10 0.6015595 0.060 0.160 2.056947e-05 2
## AL138724.1 5.620008e-10 -0.4054342 0.009 0.073 2.057035e-05 2
## RTF1 5.645266e-10 0.7661185 0.159 0.326 2.066280e-05 2
## ZNF101.1 5.652350e-10 0.4364427 0.060 0.160 2.068873e-05 2
## CMTM8.2 5.652744e-10 0.1650346 0.026 0.102 2.069017e-05 2
## RNF168.2 5.657311e-10 0.5474910 0.078 0.190 2.070689e-05 2
## MIS18BP1 5.771664e-10 0.9302894 0.188 0.377 2.112544e-05 2
## DPEP2.1 5.824835e-10 0.1706037 0.047 0.137 2.132006e-05 2
## SRXN1 5.945186e-10 -1.2780201 0.006 0.067 2.176057e-05 2
## RAB5A.1 5.956697e-10 0.7227333 0.138 0.290 2.180270e-05 2
## PRR5.2 5.963698e-10 0.7450370 0.066 0.170 2.182833e-05 2
## GTF3C2.1 5.968080e-10 0.2808354 0.038 0.123 2.184437e-05 2
## CCDC71L.1 5.989930e-10 -0.6277159 0.018 0.089 2.192434e-05 2
## NAGPA 6.007567e-10 0.2106258 0.023 0.097 2.198890e-05 2
## SLK.1 6.014006e-10 0.6717031 0.128 0.271 2.201246e-05 2
## TADA1 6.049554e-10 -0.5509336 0.018 0.089 2.214258e-05 2
## OXCT1.2 6.086598e-10 0.8592006 0.138 0.290 2.227817e-05 2
## ANKRD54 6.133657e-10 0.3437475 0.023 0.097 2.245041e-05 2
## METAP1.1 6.151472e-10 0.4703540 0.071 0.177 2.251562e-05 2
## LINC00861.2 6.185522e-10 0.5536012 0.119 0.255 2.264025e-05 2
## HJURP.2 6.233418e-10 -2.1791853 0.003 0.061 2.281556e-05 2
## COQ2.1 6.303117e-10 -0.2409359 0.012 0.078 2.307067e-05 2
## URI1.1 6.348026e-10 0.6943605 0.241 0.474 2.323504e-05 2
## OOEP 6.412762e-10 -6.0589150 0.000 0.055 2.347199e-05 2
## DHX15.1 6.454925e-10 0.8693515 0.122 0.263 2.362631e-05 2
## TELO2.2 6.487439e-10 0.4365460 0.027 0.105 2.374532e-05 2
## DERA 6.524111e-10 0.2841007 0.039 0.125 2.387955e-05 2
## UBXN2A.1 6.542286e-10 0.8193778 0.051 0.145 2.394608e-05 2
## SLC33A1 6.595068e-10 0.4185627 0.048 0.140 2.413927e-05 2
## ATP23.1 6.676471e-10 0.1880121 0.018 0.089 2.443722e-05 2
## SLC25A33 6.683660e-10 0.6899134 0.042 0.130 2.446353e-05 2
## KIF2C.2 6.723119e-10 -1.5372000 0.003 0.061 2.460796e-05 2
## CAND1.1 6.736425e-10 0.5845839 0.099 0.223 2.465666e-05 2
## CDK10 6.743527e-10 0.5386812 0.045 0.135 2.468266e-05 2
## EIF4EBP3.1 6.886035e-10 -1.2306065 0.006 0.067 2.520427e-05 2
## KLRB1.2 6.910901e-10 -0.8962037 0.063 0.157 2.529528e-05 2
## PHTF1 6.949436e-10 0.7004939 0.065 0.167 2.543632e-05 2
## RAD50 6.976328e-10 0.8961137 0.129 0.274 2.553476e-05 2
## GANAB 6.997676e-10 0.5118255 0.107 0.235 2.561290e-05 2
## ESCO1 7.065163e-10 0.7134077 0.092 0.211 2.585991e-05 2
## BCLAF1 7.066981e-10 0.6281241 0.271 0.524 2.586656e-05 2
## SEC23IP 7.084509e-10 0.2673713 0.045 0.135 2.593072e-05 2
## EXOC3 7.348182e-10 0.4772406 0.059 0.157 2.689582e-05 2
## MPND.1 7.402028e-10 -0.2284643 0.014 0.081 2.709290e-05 2
## ZC3H7A.1 7.402370e-10 0.8143743 0.098 0.221 2.709415e-05 2
## NAA50 7.454391e-10 0.6826699 0.125 0.267 2.728456e-05 2
## RNF11.1 7.466113e-10 0.6061049 0.057 0.155 2.732747e-05 2
## MUS81 7.472454e-10 0.8390511 0.071 0.177 2.735068e-05 2
## STK17B.1 7.565724e-10 1.1580504 0.535 0.684 2.769206e-05 2
## ST6GALNAC6.1 7.583804e-10 0.6618380 0.077 0.187 2.775824e-05 2
## RAMP1.1 7.664432e-10 -0.8805163 0.017 0.086 2.805335e-05 2
## MKI67.2 7.759658e-10 -0.3603213 0.066 0.163 2.840190e-05 2
## SFMBT2.2 7.852759e-10 2.5043442 0.239 0.178 2.874267e-05 2
## ASPM.2 7.876767e-10 -0.7132404 0.021 0.093 2.883054e-05 2
## ARHGAP19.2 7.886731e-10 0.4458763 0.033 0.114 2.886701e-05 2
## GEMIN4.2 7.901469e-10 -1.4183526 0.006 0.066 2.892096e-05 2
## CCDC127 7.957821e-10 -0.5835911 0.014 0.080 2.912721e-05 2
## MTRES1.1 8.000490e-10 0.1391197 0.024 0.099 2.928339e-05 2
## LRRC14 8.085566e-10 -0.3514973 0.021 0.094 2.959479e-05 2
## NEIL3.2 8.194428e-10 -0.5419216 0.012 0.077 2.999325e-05 2
## ANKRD13D 8.210066e-10 1.0099800 0.137 0.288 3.005048e-05 2
## CYTOR.1 8.228703e-10 0.8489916 0.174 0.350 3.011870e-05 2
## ETFDH 8.249504e-10 0.4161388 0.035 0.117 3.019483e-05 2
## SMIM4 8.266640e-10 -0.1931860 0.018 0.088 3.025756e-05 2
## LDOC1 8.519550e-10 -0.4348255 0.011 0.075 3.118326e-05 2
## AC012146.1 8.773305e-10 -1.4017531 0.005 0.063 3.211205e-05 2
## GALNT1.1 8.865926e-10 1.0540490 0.068 0.172 3.245106e-05 2
## INCENP.2 8.916762e-10 -0.6820415 0.012 0.077 3.263713e-05 2
## C17orf75.1 8.927178e-10 0.4652510 0.036 0.119 3.267526e-05 2
## WSB1.1 9.000151e-10 0.9571536 0.171 0.345 3.294235e-05 2
## POLR2M 9.086873e-10 -0.2539447 0.024 0.098 3.325977e-05 2
## IBTK 9.100787e-10 0.7723416 0.110 0.240 3.331070e-05 2
## DTWD1 9.176996e-10 0.4284654 0.033 0.114 3.358964e-05 2
## HSD17B4.1 9.264976e-10 0.5969756 0.062 0.161 3.391167e-05 2
## YIPF6 9.361414e-10 -0.2646210 0.012 0.077 3.426465e-05 2
## SNAP47.1 9.397442e-10 0.6199940 0.065 0.166 3.439652e-05 2
## CYP20A1.1 9.468829e-10 0.7063636 0.095 0.215 3.465781e-05 2
## SLFN12L 9.800184e-10 2.0526979 0.386 0.398 3.587063e-05 2
## SLC25A32 1.005078e-09 0.6884615 0.062 0.161 3.678785e-05 2
## CD46.2 1.020986e-09 0.7305120 0.209 0.412 3.737014e-05 2
## YIPF1.1 1.033880e-09 -0.3877702 0.015 0.082 3.784209e-05 2
## MAN1B1.1 1.035413e-09 0.7048624 0.065 0.166 3.789819e-05 2
## ANLN.2 1.035724e-09 -5.1267634 0.002 0.056 3.790957e-05 2
## BLM.2 1.041380e-09 0.6026684 0.047 0.136 3.811657e-05 2
## BRD3.1 1.044958e-09 0.4380846 0.041 0.126 3.824753e-05 2
## QRICH1 1.051205e-09 0.7431285 0.104 0.230 3.847620e-05 2
## DTNBP1 1.063892e-09 0.6944712 0.078 0.188 3.894059e-05 2
## STAMBP 1.065533e-09 0.3989846 0.050 0.141 3.900065e-05 2
## OSER1.1 1.073416e-09 0.3628745 0.078 0.188 3.928919e-05 2
## ZC3H8 1.074841e-09 0.6188327 0.050 0.141 3.934133e-05 2
## SPRYD3.1 1.075645e-09 -0.1340861 0.026 0.100 3.937078e-05 2
## TMEM214 1.078583e-09 0.3573403 0.050 0.141 3.947830e-05 2
## C15orf39 1.085979e-09 0.2422232 0.033 0.114 3.974900e-05 2
## CENPL.2 1.086793e-09 -0.1632119 0.012 0.077 3.977881e-05 2
## DNAJB14.1 1.092081e-09 0.8933753 0.164 0.332 3.997236e-05 2
## RCN1 1.096533e-09 -0.2982644 0.023 0.095 4.013528e-05 2
## PPP4R3A.1 1.097043e-09 0.8251731 0.168 0.339 4.015395e-05 2
## FAM122B 1.111569e-09 0.5904650 0.048 0.138 4.068565e-05 2
## SAMD9.2 1.114267e-09 0.8947371 0.108 0.237 4.078439e-05 2
## NT5C3A 1.117803e-09 0.7128462 0.110 0.238 4.091382e-05 2
## CCDC34.2 1.135363e-09 -1.3260080 0.009 0.071 4.155656e-05 2
## HDGFL3 1.168255e-09 0.6176245 0.075 0.182 4.276046e-05 2
## TMEM263 1.172666e-09 0.5171748 0.059 0.156 4.292191e-05 2
## REXO4 1.174577e-09 0.1060890 0.018 0.087 4.299186e-05 2
## CDK5RAP1 1.196291e-09 0.5428982 0.044 0.131 4.378665e-05 2
## BTBD1 1.204729e-09 0.7032156 0.102 0.227 4.409551e-05 2
## AC013264.1 1.207478e-09 -0.4411788 0.011 0.074 4.419610e-05 2
## NR2F6.1 1.211982e-09 -0.6651601 0.009 0.071 4.436096e-05 2
## RRP8 1.221790e-09 0.2024360 0.024 0.098 4.471996e-05 2
## POLB 1.222587e-09 0.3196705 0.041 0.126 4.474913e-05 2
## TIPIN.2 1.243778e-09 0.5113635 0.029 0.106 4.552475e-05 2
## DDX52 1.247983e-09 0.7144588 0.036 0.118 4.567869e-05 2
## CD38.2 1.252806e-09 0.7167747 0.125 0.261 4.585520e-05 2
## ARFIP1 1.271152e-09 0.6773069 0.072 0.177 4.652670e-05 2
## ABCD4 1.275531e-09 0.2934278 0.041 0.126 4.668698e-05 2
## WDR48 1.297903e-09 0.6943718 0.065 0.165 4.750583e-05 2
## ANKRA2 1.307314e-09 -0.1947126 0.024 0.097 4.785032e-05 2
## DYRK2.1 1.337957e-09 0.3178480 0.066 0.167 4.897191e-05 2
## AP3M1 1.343846e-09 1.0281013 0.068 0.170 4.918744e-05 2
## STIL.2 1.345497e-09 -1.1006277 0.008 0.068 4.924788e-05 2
## CASP3 1.352158e-09 0.6481665 0.077 0.184 4.949169e-05 2
## WDR12.1 1.352496e-09 0.6867053 0.045 0.133 4.950404e-05 2
## LDB1 1.362014e-09 0.7286330 0.096 0.217 4.985243e-05 2
## TRIP13.2 1.369777e-09 -1.6159511 0.003 0.059 5.013657e-05 2
## PERP.2 1.372070e-09 0.5690403 0.042 0.128 5.022050e-05 2
## NOS3.1 1.376582e-09 -0.8372032 0.009 0.071 5.038567e-05 2
## NUDT8.1 1.381403e-09 -1.2361295 0.006 0.065 5.056210e-05 2
## LINC01003 1.413396e-09 -0.6046998 0.009 0.071 5.173312e-05 2
## OMA1 1.415393e-09 -0.4524616 0.021 0.092 5.180622e-05 2
## UBE2G2 1.430386e-09 0.7232015 0.119 0.254 5.235501e-05 2
## CKAP2L.2 1.437487e-09 -1.2314798 0.005 0.062 5.261491e-05 2
## BUB1.2 1.444801e-09 -0.4129382 0.015 0.081 5.288261e-05 2
## GATA3 1.445132e-09 0.7174812 0.229 0.439 5.289474e-05 2
## SLC25A23 1.451971e-09 -0.2249823 0.021 0.092 5.314504e-05 2
## HIC1.2 1.467582e-09 0.6831020 0.075 0.182 5.371643e-05 2
## PGAM5.1 1.491615e-09 -0.3263446 0.020 0.089 5.459610e-05 2
## BEX3.1 1.503996e-09 -0.6066264 0.009 0.070 5.504927e-05 2
## AP2A1.1 1.527513e-09 0.3975756 0.053 0.145 5.591001e-05 2
## FMNL1.2 1.527913e-09 0.7690173 0.232 0.451 5.592467e-05 2
## PITPNB 1.554349e-09 0.7043201 0.120 0.256 5.689229e-05 2
## ABRAXAS2 1.560784e-09 0.1774635 0.039 0.122 5.712781e-05 2
## JMJD4.2 1.571591e-09 0.3337095 0.029 0.105 5.752339e-05 2
## IST1 1.583296e-09 0.8038980 0.119 0.254 5.795181e-05 2
## PHC2 1.598345e-09 0.3261708 0.057 0.152 5.850264e-05 2
## C7orf26 1.601459e-09 0.3845555 0.029 0.105 5.861659e-05 2
## PNPT1 1.622864e-09 0.6348255 0.054 0.147 5.940008e-05 2
## CHAC2.2 1.638368e-09 -1.6773288 0.005 0.061 5.996755e-05 2
## LIAS 1.638777e-09 -0.2886640 0.012 0.076 5.998250e-05 2
## MYBBP1A.1 1.640168e-09 0.3706759 0.035 0.115 6.003343e-05 2
## AC010642.2 1.652327e-09 0.1205578 0.017 0.084 6.047847e-05 2
## TMEM165 1.663498e-09 0.9008700 0.123 0.261 6.088735e-05 2
## CAAP1 1.666102e-09 0.6809157 0.083 0.194 6.098266e-05 2
## NCLN.1 1.676813e-09 0.5541186 0.045 0.132 6.137471e-05 2
## CWF19L1 1.699144e-09 0.3445005 0.048 0.137 6.219209e-05 2
## RAD51.2 1.707541e-09 -0.2554960 0.017 0.084 6.249942e-05 2
## PPP1R3F.1 1.711490e-09 -0.3027437 0.026 0.099 6.264395e-05 2
## USP8 1.722332e-09 0.8834752 0.126 0.266 6.304080e-05 2
## CDC7.2 1.808756e-09 -1.2748853 0.009 0.070 6.620409e-05 2
## ARMH1.2 1.812455e-09 0.5049537 0.062 0.159 6.633949e-05 2
## GIMAP8.1 1.813767e-09 0.3805445 0.053 0.145 6.638752e-05 2
## RXYLT1.1 1.820080e-09 -0.1924521 0.020 0.089 6.661855e-05 2
## RRP15 1.829592e-09 -0.4056618 0.018 0.086 6.696674e-05 2
## RAB10 1.833656e-09 0.9547350 0.195 0.389 6.711549e-05 2
## GABPB1-IT1 1.834579e-09 0.3998703 0.069 0.171 6.714926e-05 2
## AP5Z1 1.838372e-09 0.8002372 0.063 0.162 6.728810e-05 2
## ADPGK 1.882471e-09 0.8558364 0.095 0.213 6.890221e-05 2
## MAP11 1.891115e-09 0.2070033 0.027 0.102 6.921858e-05 2
## TUT1.1 1.919943e-09 0.1302539 0.024 0.097 7.027376e-05 2
## CENPE.2 1.944232e-09 -0.1156093 0.023 0.094 7.116278e-05 2
## CEP41.1 1.955306e-09 -1.6326952 0.003 0.058 7.156810e-05 2
## BNIP1.1 1.961705e-09 0.2036182 0.014 0.078 7.180233e-05 2
## PTCD3.1 1.965002e-09 0.7152829 0.072 0.176 7.192300e-05 2
## UBE2Z 1.983652e-09 0.6039262 0.098 0.217 7.260564e-05 2
## PREX1.1 2.005172e-09 2.0331719 0.414 0.452 7.339329e-05 2
## NAT10 2.013295e-09 0.4209521 0.054 0.146 7.369062e-05 2
## XPC.2 2.025429e-09 0.7606318 0.164 0.326 7.413475e-05 2
## GTF2H4 2.027430e-09 -0.7166445 0.012 0.075 7.420800e-05 2
## PHF20L1 2.047122e-09 0.6986947 0.138 0.285 7.492875e-05 2
## ELP2 2.047346e-09 0.6242164 0.087 0.200 7.493696e-05 2
## SRFBP1 2.069400e-09 0.6652841 0.060 0.156 7.574419e-05 2
## GIT1 2.072712e-09 -0.2865162 0.018 0.086 7.586539e-05 2
## SRD5A3.1 2.080260e-09 -0.2803410 0.011 0.072 7.614167e-05 2
## MTF2 2.105046e-09 0.6371839 0.081 0.190 7.704888e-05 2
## RNF4.1 2.113914e-09 0.7435019 0.128 0.267 7.737348e-05 2
## SCAMP4 2.120604e-09 -0.1494766 0.020 0.088 7.761833e-05 2
## TOPORS 2.126087e-09 0.5736735 0.039 0.122 7.781905e-05 2
## ZC3H14 2.144005e-09 0.7463475 0.059 0.153 7.847486e-05 2
## IMPAD1 2.144338e-09 0.5171637 0.060 0.156 7.848704e-05 2
## SLC25A36 2.151833e-09 0.8653744 0.069 0.171 7.876139e-05 2
## MYL5 2.154983e-09 -1.0150289 0.008 0.067 7.887670e-05 2
## NAA60 2.192769e-09 0.2201908 0.033 0.111 8.025973e-05 2
## IFIH1.1 2.205509e-09 0.3332470 0.056 0.148 8.072604e-05 2
## PLAAT3.2 2.214046e-09 -0.6457115 0.014 0.078 8.103851e-05 2
## SAMD1.2 2.258012e-09 0.6152575 0.033 0.111 8.264777e-05 2
## PNKP.1 2.294807e-09 0.9869214 0.096 0.215 8.399451e-05 2
## TMEM154 2.303337e-09 0.7194030 0.036 0.116 8.430673e-05 2
## NQO2.1 2.370489e-09 1.1504181 0.047 0.134 8.676463e-05 2
## COX10 2.396731e-09 0.7194720 0.056 0.148 8.772516e-05 2
## DCUN1D1.1 2.399535e-09 0.6526385 0.068 0.168 8.782776e-05 2
## SLAIN2.1 2.410891e-09 0.7014574 0.120 0.254 8.824344e-05 2
## TMEM129 2.415460e-09 0.1486575 0.021 0.091 8.841067e-05 2
## EEF2KMT.1 2.416529e-09 -0.4846976 0.011 0.072 8.844978e-05 2
## EXOSC10 2.424178e-09 0.6466011 0.087 0.200 8.872975e-05 2
## IFT22.1 2.434711e-09 -0.7372368 0.009 0.069 8.911528e-05 2
## ZNF770 2.435091e-09 0.2567448 0.060 0.155 8.912921e-05 2
## SECISBP2L.1 2.443474e-09 0.4603197 0.093 0.208 8.943602e-05 2
## DENND1C.1 2.455550e-09 0.8609374 0.162 0.327 8.987805e-05 2
## IKBKE 2.484183e-09 1.0333840 0.102 0.225 9.092605e-05 2
## CCNF.2 2.537905e-09 -0.9448738 0.009 0.069 9.289240e-05 2
## PDXDC1.1 2.538543e-09 0.9209408 0.113 0.241 9.291576e-05 2
## RASSF2 2.547976e-09 0.5548725 0.062 0.158 9.326103e-05 2
## DHX30.1 2.576544e-09 0.9516099 0.084 0.195 9.430668e-05 2
## DARS2 2.604098e-09 -0.6373003 0.009 0.069 9.531519e-05 2
## POGK 2.610348e-09 0.1113379 0.029 0.103 9.554395e-05 2
## IL2RB.1 2.614248e-09 0.6234379 0.161 0.318 9.568671e-05 2
## TIGAR.1 2.614271e-09 0.5258867 0.069 0.170 9.568756e-05 2
## MMUT.1 2.626493e-09 0.2233284 0.021 0.090 9.613489e-05 2
## CEP104 2.631320e-09 0.6839195 0.056 0.148 9.631158e-05 2
## CIAO3 2.663108e-09 0.1074210 0.024 0.095 9.747509e-05 2
## MED25 2.743163e-09 0.7519268 0.041 0.123 1.004052e-04 2
## KAT2A.1 2.762399e-09 0.2383295 0.014 0.077 1.011093e-04 2
## LSG1.1 2.798639e-09 0.5377279 0.047 0.133 1.024358e-04 2
## CDC23.1 2.824099e-09 0.1643761 0.023 0.093 1.033677e-04 2
## HSBP1L1 2.829720e-09 -0.6043732 0.009 0.069 1.035734e-04 2
## KCTD10 2.842240e-09 -0.7091750 0.014 0.077 1.040317e-04 2
## CENPN.2 2.846134e-09 0.6311871 0.027 0.100 1.041742e-04 2
## GOSR1 2.848583e-09 0.7776409 0.098 0.216 1.042638e-04 2
## RNF115 2.872747e-09 0.7873536 0.116 0.246 1.051483e-04 2
## CLK4 2.876713e-09 0.2231569 0.050 0.138 1.052935e-04 2
## ARL15.1 2.880192e-09 2.3737295 0.272 0.224 1.054208e-04 2
## TRIM4 2.881171e-09 0.5768221 0.074 0.177 1.054566e-04 2
## HAUS6.1 2.914535e-09 0.5978995 0.054 0.145 1.066778e-04 2
## ING5.1 2.932392e-09 0.4177126 0.039 0.121 1.073314e-04 2
## GCFC2 2.951219e-09 0.1573851 0.033 0.111 1.080205e-04 2
## LAS1L.1 2.956640e-09 1.0799800 0.050 0.138 1.082189e-04 2
## ELMO1.2 2.956739e-09 1.8620844 0.406 0.439 1.082226e-04 2
## AC093157.1.2 2.961191e-09 0.5659729 0.051 0.140 1.083855e-04 2
## TMEM159.1 3.021430e-09 -0.5797282 0.014 0.077 1.105904e-04 2
## BEX2.1 3.022286e-09 -3.6593741 0.003 0.057 1.106217e-04 2
## IP6K2 3.030725e-09 0.8001350 0.104 0.227 1.109306e-04 2
## RFK 3.054186e-09 -0.1148934 0.030 0.105 1.117893e-04 2
## AAR2.1 3.068444e-09 0.2489196 0.030 0.105 1.123112e-04 2
## CHD4 3.113515e-09 0.8146494 0.150 0.304 1.139609e-04 2
## ALDH3A2 3.208194e-09 -1.0470353 0.012 0.074 1.174263e-04 2
## DUSP28 3.211616e-09 -0.2397050 0.021 0.090 1.175516e-04 2
## TRAF2.2 3.252775e-09 0.6343736 0.044 0.128 1.190581e-04 2
## ARRDC2 3.255126e-09 0.7712333 0.068 0.167 1.191441e-04 2
## PMS1 3.345417e-09 0.8315718 0.086 0.196 1.224490e-04 2
## CEP19.2 3.351186e-09 0.3331788 0.027 0.100 1.226601e-04 2
## SURF6 3.364662e-09 0.4697834 0.044 0.128 1.231534e-04 2
## JPT2.2 3.433603e-09 0.3467991 0.033 0.110 1.256767e-04 2
## PDCD2L 3.464796e-09 -1.5077142 0.006 0.062 1.268185e-04 2
## NXT2 3.476270e-09 -1.5491759 0.006 0.062 1.272384e-04 2
## GGA1.1 3.498263e-09 0.6544300 0.086 0.195 1.280434e-04 2
## AC016831.7.1 3.534741e-09 2.7369101 0.140 0.081 1.293786e-04 2
## SNX4.1 3.553521e-09 0.6221046 0.068 0.166 1.300660e-04 2
## SAMD10.1 3.561683e-09 -0.1341977 0.020 0.087 1.303647e-04 2
## HSH2D.1 3.581886e-09 0.8502080 0.053 0.142 1.311042e-04 2
## BPNT1 3.583535e-09 -0.4513024 0.015 0.079 1.311646e-04 2
## NUDCD1 3.588317e-09 0.6076447 0.048 0.135 1.313396e-04 2
## ZNF433-AS1 3.594956e-09 0.7009904 0.047 0.132 1.315826e-04 2
## MAIP1 3.600233e-09 -0.2295691 0.020 0.087 1.317757e-04 2
## HLTF.1 3.607104e-09 0.8864061 0.042 0.125 1.320272e-04 2
## MAP2K1.1 3.615713e-09 0.8846094 0.170 0.337 1.323423e-04 2
## ZFYVE27 3.637255e-09 0.5444923 0.032 0.107 1.331308e-04 2
## NDC1.2 3.684703e-09 0.2402885 0.023 0.092 1.348675e-04 2
## DHRS4-AS1 3.725767e-09 -0.3001000 0.015 0.079 1.363705e-04 2
## GSTZ1.1 3.746486e-09 -0.4866075 0.009 0.068 1.371289e-04 2
## GLI4 3.796279e-09 0.2223286 0.029 0.102 1.389514e-04 2
## TNKS2 3.806434e-09 0.7211363 0.150 0.303 1.393231e-04 2
## TSPAN32.2 3.861479e-09 0.7538800 0.080 0.185 1.413378e-04 2
## TAF1B 3.917000e-09 0.6817343 0.069 0.168 1.433700e-04 2
## DYNC1LI2 3.937105e-09 0.8965847 0.074 0.176 1.441059e-04 2
## SMAD2 3.940400e-09 0.7822780 0.180 0.354 1.442265e-04 2
## CHAF1B.2 4.034004e-09 0.2030892 0.012 0.073 1.476526e-04 2
## OAZ2.1 4.081100e-09 -0.1669266 0.023 0.092 1.493764e-04 2
## ELK4 4.099518e-09 0.6344676 0.095 0.210 1.500506e-04 2
## PARP3.1 4.163470e-09 -0.3397142 0.017 0.081 1.523913e-04 2
## TSR1.2 4.212582e-09 0.6385091 0.039 0.119 1.541889e-04 2
## ZNF106 4.272929e-09 0.8488609 0.117 0.246 1.563977e-04 2
## SLTM 4.286959e-09 0.7529183 0.229 0.440 1.569113e-04 2
## SOCS1.1 4.357644e-09 0.8573048 0.170 0.336 1.594985e-04 2
## PRKAA1 4.359063e-09 0.8087006 0.096 0.213 1.595504e-04 2
## PPP3CA.2 4.443419e-09 2.2569745 0.359 0.360 1.626380e-04 2
## EXOC6 4.472805e-09 0.6187783 0.072 0.173 1.637136e-04 2
## KATNB1.1 4.485936e-09 -0.2127496 0.024 0.094 1.641942e-04 2
## ZNF721 4.506999e-09 0.9229442 0.104 0.225 1.649652e-04 2
## ESRRA.1 4.517555e-09 0.8707544 0.050 0.137 1.653515e-04 2
## ZNF236-DT 4.522630e-09 -0.9770683 0.009 0.067 1.655373e-04 2
## INTS12 4.536861e-09 0.2304621 0.036 0.114 1.660582e-04 2
## PNPO 4.609167e-09 -0.2294571 0.009 0.067 1.687047e-04 2
## SFT2D3 4.630174e-09 -1.3623782 0.008 0.064 1.694736e-04 2
## MBD3.2 4.646499e-09 0.4529257 0.027 0.099 1.700711e-04 2
## MAP4K5 4.646512e-09 0.9128312 0.095 0.210 1.700716e-04 2
## MGAT5.2 4.647113e-09 2.3438114 0.238 0.181 1.700936e-04 2
## NBEAL1 4.651806e-09 0.5050682 0.065 0.161 1.702654e-04 2
## NOL8 4.654604e-09 0.9503424 0.096 0.212 1.703678e-04 2
## C12orf29 4.711222e-09 -1.0726737 0.015 0.078 1.724402e-04 2
## METTL18 4.770519e-09 -1.2048281 0.009 0.067 1.746105e-04 2
## TOB1.1 4.787486e-09 0.8876887 0.114 0.241 1.752316e-04 2
## RRAGD.2 4.801983e-09 -1.1168772 0.005 0.059 1.757622e-04 2
## FOXRED1.1 4.805304e-09 0.8928176 0.041 0.122 1.758837e-04 2
## CDC27.1 4.829689e-09 0.8775206 0.125 0.259 1.767763e-04 2
## CHCHD3.1 4.862885e-09 1.0009790 0.135 0.277 1.779913e-04 2
## COG4 4.876148e-09 0.8775614 0.075 0.178 1.784768e-04 2
## UBE2D4 4.899455e-09 -1.3619956 0.009 0.067 1.793299e-04 2
## ITGB1.2 4.917566e-09 0.4983965 0.155 0.302 1.799927e-04 2
## ING1 4.954527e-09 0.7104752 0.045 0.129 1.813456e-04 2
## ZNF217.2 5.006333e-09 0.7978524 0.205 0.398 1.832418e-04 2
## GALE.1 5.009222e-09 -0.4914092 0.011 0.070 1.833475e-04 2
## SCLY.1 5.021646e-09 0.4272549 0.044 0.126 1.838023e-04 2
## SREK1IP1 5.028752e-09 0.6202638 0.069 0.168 1.840624e-04 2
## SMG6.1 5.176626e-09 2.0006799 0.343 0.328 1.894749e-04 2
## HCFC1.2 5.198701e-09 0.7852256 0.075 0.178 1.902829e-04 2
## ASCC1 5.258956e-09 0.1755198 0.038 0.116 1.924883e-04 2
## TRGV10.2 5.276189e-09 -0.3026622 0.035 0.111 1.931191e-04 2
## STARD10.1 5.287833e-09 -1.3033819 0.003 0.055 1.935452e-04 2
## ACOT9 5.294039e-09 0.5391760 0.036 0.114 1.937724e-04 2
## RAB35.1 5.304608e-09 0.7110682 0.071 0.170 1.941593e-04 2
## RMDN3 5.329662e-09 0.4530339 0.050 0.136 1.950763e-04 2
## TMEM127 5.369556e-09 0.2154808 0.045 0.128 1.965365e-04 2
## LINC00847 5.417161e-09 -0.5606937 0.017 0.080 1.982789e-04 2
## FAM98A 5.425403e-09 0.3042274 0.032 0.106 1.985806e-04 2
## TBL2 5.429701e-09 -0.2158737 0.015 0.078 1.987379e-04 2
## USO1 5.477691e-09 0.8857818 0.146 0.294 2.004944e-04 2
## BTN3A1.1 5.501343e-09 0.7592735 0.114 0.241 2.013602e-04 2
## TKFC.1 5.524561e-09 -0.2910302 0.011 0.070 2.022100e-04 2
## HGH1 5.524591e-09 -0.1133244 0.018 0.083 2.022111e-04 2
## LIPE 5.543565e-09 -0.2668637 0.011 0.070 2.029056e-04 2
## ZMYM5 5.658533e-09 0.7185002 0.069 0.167 2.071136e-04 2
## COPA.1 5.661529e-09 0.8791811 0.165 0.327 2.072233e-04 2
## TRIB2 5.765497e-09 0.2694535 0.041 0.121 2.110287e-04 2
## CCPG1.1 5.782548e-09 0.5718412 0.075 0.177 2.116528e-04 2
## C6orf47 5.812060e-09 0.1179009 0.029 0.101 2.127330e-04 2
## ZFR 5.836985e-09 0.8521529 0.176 0.345 2.136453e-04 2
## FANCD2.2 5.884026e-09 -0.2198526 0.024 0.093 2.153671e-04 2
## RBM25 5.894463e-09 0.8148677 0.260 0.500 2.157491e-04 2
## CDC20.2 5.909018e-09 -1.2870504 0.005 0.058 2.162819e-04 2
## UBE2G1.1 6.023037e-09 0.8084520 0.159 0.317 2.204552e-04 2
## RBM14.1 6.031852e-09 0.6744681 0.108 0.231 2.207779e-04 2
## FAM50B.1 6.065823e-09 -0.5628955 0.008 0.064 2.220212e-04 2
## OAS3.2 6.071672e-09 -0.1544019 0.026 0.096 2.222353e-04 2
## CHERP 6.100039e-09 0.6778709 0.053 0.140 2.232736e-04 2
## IL18BP 6.239147e-09 -0.1745642 0.015 0.077 2.283653e-04 2
## LINC00324.2 6.306939e-09 0.6412215 0.026 0.096 2.308466e-04 2
## ALKBH3.1 6.389516e-09 0.6067184 0.044 0.126 2.338690e-04 2
## CCR4.2 6.413485e-09 0.5234126 0.051 0.137 2.347464e-04 2
## RMDN1 6.439247e-09 0.6918102 0.069 0.167 2.356893e-04 2
## CLN5.2 6.587225e-09 0.1792381 0.023 0.090 2.411056e-04 2
## MTERF3.1 6.634643e-09 0.2341044 0.027 0.098 2.428412e-04 2
## MTFR1L.1 6.705315e-09 0.5798516 0.030 0.103 2.454279e-04 2
## TRMT2A 6.766508e-09 0.4853548 0.047 0.130 2.476677e-04 2
## TXLNG 6.778538e-09 0.2477224 0.032 0.105 2.481080e-04 2
## RNF20 6.837984e-09 0.3774809 0.057 0.147 2.502839e-04 2
## SAP30L 6.862261e-09 0.1932954 0.038 0.115 2.511725e-04 2
## CIT.2 6.923409e-09 -1.1489158 0.009 0.066 2.534106e-04 2
## OLFM2 6.944799e-09 -0.7708899 0.009 0.066 2.541935e-04 2
## CCDC18.2 6.964791e-09 0.7690203 0.054 0.142 2.549253e-04 2
## GTPBP3.1 7.052988e-09 -0.2232517 0.017 0.080 2.581535e-04 2
## USP22.2 7.080858e-09 0.7439431 0.098 0.212 2.591736e-04 2
## PIAS4 7.090478e-09 0.3567349 0.026 0.095 2.595257e-04 2
## CWC22.1 7.151518e-09 0.4873900 0.057 0.147 2.617599e-04 2
## BX890604.2 7.230664e-09 0.1018352 0.018 0.082 2.646568e-04 2
## ZNF880 7.440894e-09 -0.5436855 0.009 0.066 2.723516e-04 2
## LIPT1 7.494513e-09 -2.6472240 0.002 0.051 2.743142e-04 2
## TMC6.1 7.643082e-09 0.9046131 0.229 0.439 2.797521e-04 2
## SH2B1 7.643667e-09 0.5305542 0.042 0.123 2.797735e-04 2
## LACTB.1 7.690857e-09 0.4296433 0.033 0.108 2.815007e-04 2
## TRIM5 7.775934e-09 0.5497390 0.032 0.105 2.846148e-04 2
## SPAG16.2 7.797410e-09 -0.2927072 0.021 0.087 2.854008e-04 2
## JOSD1.2 7.827555e-09 0.9937859 0.059 0.149 2.865042e-04 2
## NBR1 7.838112e-09 0.6909289 0.122 0.251 2.868906e-04 2
## MAGEF1.1 7.856445e-09 -1.6464186 0.008 0.063 2.875616e-04 2
## XPOT.2 7.909567e-09 0.5978455 0.068 0.164 2.895060e-04 2
## RAB3GAP1.2 7.962493e-09 0.7851691 0.060 0.152 2.914432e-04 2
## ESCO2.2 7.966661e-09 -0.3054975 0.009 0.066 2.915957e-04 2
## ECHDC2.2 7.996424e-09 0.2138777 0.030 0.102 2.926851e-04 2
## ZFYVE21.1 8.200585e-09 -0.2504717 0.015 0.077 3.001578e-04 2
## NIPAL3 8.202122e-09 0.5982363 0.089 0.198 3.002141e-04 2
## LFNG.1 8.270842e-09 0.9168380 0.093 0.204 3.027294e-04 2
## DDX1 8.283629e-09 0.4531320 0.048 0.132 3.031974e-04 2
## RSBN1 8.296961e-09 0.8507523 0.093 0.205 3.036854e-04 2
## TMEM107.1 8.304318e-09 1.0132221 0.120 0.249 3.039546e-04 2
## SUV39H1.2 8.398516e-09 0.2923921 0.011 0.068 3.074025e-04 2
## SPDL1.2 8.557942e-09 -2.3277723 0.003 0.054 3.132378e-04 2
## SAFB2 8.663208e-09 0.8834709 0.123 0.254 3.170908e-04 2
## PXN-AS1.1 8.744720e-09 -0.4251451 0.014 0.074 3.200742e-04 2
## HBS1L.1 8.791925e-09 0.9271175 0.129 0.264 3.218020e-04 2
## TTI2.1 8.802295e-09 0.2401643 0.023 0.090 3.221816e-04 2
## UBXN6 9.084741e-09 -0.7276078 0.005 0.057 3.325197e-04 2
## TMEM64.1 9.136426e-09 -0.6591942 0.012 0.071 3.344115e-04 2
## DLGAP5.2 9.239451e-09 -2.3131195 0.003 0.054 3.381824e-04 2
## RPF2.1 9.459951e-09 0.7490215 0.045 0.127 3.462531e-04 2
## GIT2.1 9.626629e-09 0.8229964 0.162 0.319 3.523539e-04 2
## PINX1 9.685681e-09 0.5166728 0.038 0.115 3.545153e-04 2
## ICMT.2 9.728531e-09 0.2378400 0.041 0.119 3.560837e-04 2
## TRAF3IP2.1 9.748686e-09 -0.2490631 0.024 0.092 3.568214e-04 2
## RAD9A.1 9.749401e-09 0.9289061 0.054 0.141 3.568476e-04 2
## ZNF576.1 9.762507e-09 -0.5178807 0.009 0.065 3.573273e-04 2
## UBE3A 9.787286e-09 0.9403748 0.203 0.393 3.582342e-04 2
## GPR155.1 9.903438e-09 0.5971008 0.056 0.143 3.624856e-04 2
## MCM8.2 1.009218e-08 -1.7874231 0.005 0.056 3.693941e-04 2
## LUC7L3 1.024922e-08 1.0772632 0.205 0.396 3.751419e-04 2
## ESS2.1 1.035734e-08 0.4447484 0.024 0.092 3.790993e-04 2
## ZNF766 1.038839e-08 0.4768754 0.042 0.122 3.802359e-04 2
## SKP2.2 1.054990e-08 -0.1528121 0.018 0.081 3.861474e-04 2
## USP48.1 1.058907e-08 0.9231824 0.144 0.289 3.875813e-04 2
## DIS3L.1 1.072974e-08 0.6158008 0.042 0.122 3.927299e-04 2
## SMPD4.1 1.095273e-08 0.5362094 0.056 0.143 4.008918e-04 2
## KCNK6 1.096500e-08 -0.4914956 0.023 0.089 4.013410e-04 2
## TMED1 1.099380e-08 -0.6071448 0.017 0.078 4.023950e-04 2
## RCHY1 1.099716e-08 0.5133197 0.041 0.119 4.025180e-04 2
## AAMDC.1 1.104066e-08 0.2875899 0.024 0.091 4.041102e-04 2
## APPBP2 1.118020e-08 0.6115571 0.077 0.176 4.092176e-04 2
## CHST14 1.127637e-08 -0.3644122 0.012 0.070 4.127378e-04 2
## MEN1 1.128256e-08 0.2740799 0.036 0.111 4.129641e-04 2
## DBR1 1.128567e-08 -0.3828922 0.008 0.062 4.130781e-04 2
## GOLGA5 1.131101e-08 0.5976831 0.047 0.129 4.140055e-04 2
## CTNNAL1.2 1.147339e-08 -1.6011182 0.002 0.050 4.199490e-04 2
## TMEM120B 1.149420e-08 0.4693299 0.032 0.104 4.207107e-04 2
## FAM133B 1.150123e-08 0.8599053 0.110 0.231 4.209680e-04 2
## CENPB.1 1.154474e-08 0.1059017 0.018 0.081 4.225604e-04 2
## ZFPM1 1.157870e-08 0.6216192 0.051 0.136 4.238036e-04 2
## FAM219B.1 1.162050e-08 0.4289086 0.041 0.119 4.253337e-04 2
## HLA-DMB.1 1.164125e-08 -0.1335415 0.015 0.076 4.260930e-04 2
## CMTR2 1.164500e-08 -0.3689485 0.015 0.076 4.262304e-04 2
## LRRC41.1 1.167097e-08 0.8154863 0.054 0.141 4.271808e-04 2
## KLHDC4 1.178371e-08 0.6023812 0.060 0.150 4.313073e-04 2
## KIF23.2 1.180986e-08 -0.1858557 0.012 0.070 4.322644e-04 2
## CREBL2 1.181567e-08 0.2386368 0.038 0.114 4.324770e-04 2
## USP28.1 1.190343e-08 0.6463497 0.059 0.148 4.356893e-04 2
## DBNDD2.2 1.195132e-08 -2.1552920 0.005 0.056 4.374423e-04 2
## GTSE1.2 1.196930e-08 -0.4096845 0.012 0.070 4.381002e-04 2
## C11orf24.1 1.204099e-08 -0.6840421 0.005 0.056 4.407244e-04 2
## LIMS1 1.206618e-08 0.7566877 0.159 0.312 4.416464e-04 2
## TMEM140 1.208774e-08 -0.1279096 0.032 0.103 4.424355e-04 2
## SLC25A26 1.211340e-08 0.6658359 0.090 0.199 4.433746e-04 2
## SPATC1L.1 1.221521e-08 -1.4656152 0.006 0.059 4.471011e-04 2
## CHAMP1.1 1.224219e-08 0.4934856 0.047 0.128 4.480885e-04 2
## WHAMM 1.253752e-08 0.8891550 0.129 0.263 4.588984e-04 2
## DDX20 1.270563e-08 -0.2080360 0.018 0.081 4.650515e-04 2
## EXOC7 1.278744e-08 0.9150264 0.065 0.158 4.680461e-04 2
## AC245297.3 1.296159e-08 0.4697629 0.066 0.159 4.744199e-04 2
## SFR1.1 1.301210e-08 -0.2501836 0.008 0.062 4.762688e-04 2
## ZSCAN16-AS1 1.308455e-08 0.4459203 0.033 0.106 4.789207e-04 2
## HHLA3 1.325254e-08 -2.1088081 0.003 0.053 4.850696e-04 2
## USF2.1 1.342901e-08 0.6896137 0.033 0.106 4.915284e-04 2
## INIP.2 1.358650e-08 0.3826767 0.044 0.123 4.972931e-04 2
## MKRN2 1.361466e-08 0.4537551 0.035 0.108 4.983237e-04 2
## RHOH 1.362349e-08 1.3263079 0.529 0.688 4.986471e-04 2
## CPSF1 1.368236e-08 0.8382015 0.069 0.164 5.008018e-04 2
## KPNA4 1.377397e-08 0.9960446 0.110 0.231 5.041550e-04 2
## SPOP 1.401928e-08 0.7807686 0.110 0.230 5.131336e-04 2
## KHSRP.2 1.422112e-08 0.4454613 0.039 0.115 5.205215e-04 2
## YWHAZ.2 1.426127e-08 0.1566049 0.471 0.835 5.219909e-04 2
## TXK.1 1.445856e-08 0.6198376 0.083 0.185 5.292121e-04 2
## CDKN2C.2 1.449847e-08 -0.2953067 0.012 0.070 5.306731e-04 2
## RAP2A.1 1.449859e-08 0.3528215 0.036 0.111 5.306774e-04 2
## MAP3K13.1 1.465321e-08 -0.3914159 0.017 0.078 5.363368e-04 2
## PIGK 1.471268e-08 0.5140019 0.056 0.143 5.385134e-04 2
## PPP1R15A.1 1.477675e-08 1.3027842 0.535 0.678 5.408586e-04 2
## RAD54L.2 1.478916e-08 -0.7473816 0.005 0.055 5.413130e-04 2
## CLPX 1.518644e-08 0.5510695 0.071 0.167 5.558541e-04 2
## SELENOM.2 1.525469e-08 -1.4449859 0.003 0.052 5.583523e-04 2
## ZNHIT2 1.568313e-08 -0.2383244 0.012 0.069 5.740338e-04 2
## CCDC71 1.571323e-08 -0.5564207 0.015 0.075 5.751355e-04 2
## TTC3 1.579697e-08 0.9260966 0.202 0.386 5.782008e-04 2
## NPRL3.1 1.591155e-08 0.4007434 0.033 0.105 5.823947e-04 2
## KRIT1 1.610490e-08 0.7002475 0.086 0.190 5.894716e-04 2
## SLC19A1.2 1.626912e-08 0.2612637 0.021 0.085 5.954824e-04 2
## CASC3 1.630169e-08 0.7058986 0.062 0.152 5.966743e-04 2
## NPLOC4.1 1.649370e-08 0.8765472 0.068 0.161 6.037023e-04 2
## FCER1G 1.665439e-08 -4.7863931 0.002 0.049 6.095838e-04 2
## WRNIP1 1.669391e-08 0.9439486 0.108 0.228 6.110304e-04 2
## SHPRH.1 1.684089e-08 0.8357227 0.086 0.190 6.164101e-04 2
## RNF141 1.686557e-08 -0.1440447 0.023 0.087 6.173136e-04 2
## FASTKD2 1.694901e-08 0.8521326 0.057 0.145 6.203678e-04 2
## TVP23B 1.709466e-08 0.3365826 0.032 0.103 6.256987e-04 2
## LETM1 1.713206e-08 -0.1034801 0.027 0.095 6.270675e-04 2
## CKAP5.1 1.719760e-08 0.7318403 0.101 0.213 6.294667e-04 2
## REST.1 1.752697e-08 0.8451461 0.185 0.358 6.415223e-04 2
## KHK.2 1.753790e-08 -1.1248472 0.003 0.052 6.419222e-04 2
## RBBP5.1 1.771107e-08 0.4722603 0.044 0.122 6.482606e-04 2
## KLHL22.1 1.801090e-08 0.8322346 0.050 0.132 6.592350e-04 2
## KIAA1586 1.812782e-08 0.6499195 0.036 0.110 6.635143e-04 2
## TRIM52-AS1 1.817344e-08 -5.7457705 0.000 0.046 6.651844e-04 2
## ORC5 1.826816e-08 0.4938254 0.059 0.147 6.686513e-04 2
## RWDD2B 1.842438e-08 -1.0979713 0.008 0.061 6.743692e-04 2
## TMEM87B 1.848049e-08 0.8150819 0.081 0.182 6.764228e-04 2
## TSPYL1 1.848858e-08 0.8125217 0.071 0.166 6.767191e-04 2
## TRMT61A 1.850955e-08 -1.3815497 0.008 0.061 6.774867e-04 2
## SNX14.1 1.853765e-08 0.8022146 0.119 0.243 6.785150e-04 2
## PCED1A.1 1.857782e-08 -0.7299351 0.011 0.066 6.799854e-04 2
## DCAF8 1.858442e-08 0.8027377 0.120 0.247 6.802269e-04 2
## ATAD3A.2 1.867046e-08 0.3041311 0.021 0.085 6.833760e-04 2
## EEA1 1.868405e-08 0.8547203 0.102 0.217 6.838735e-04 2
## CDCA7L.2 1.871139e-08 0.5011549 0.033 0.105 6.848743e-04 2
## FANCB.2 1.874759e-08 -1.7610543 0.005 0.055 6.861994e-04 2
## PRKACB.1 1.882755e-08 0.8176269 0.218 0.415 6.891261e-04 2
## CFD 1.899115e-08 -0.3186684 0.011 0.066 6.951142e-04 2
## MTRF1L 1.913376e-08 0.1836455 0.018 0.079 7.003339e-04 2
## COIL 1.914932e-08 0.6860498 0.038 0.112 7.009034e-04 2
## POGLUT1 1.938863e-08 0.4883143 0.056 0.141 7.096626e-04 2
## SUPV3L1.1 1.940590e-08 0.7491806 0.059 0.146 7.102947e-04 2
## QRSL1.1 1.944382e-08 -0.2645819 0.017 0.077 7.116827e-04 2
## PRC1.2 1.949880e-08 0.1871265 0.029 0.097 7.136952e-04 2
## PECAM1.2 1.959028e-08 0.3007291 0.071 0.164 7.170434e-04 2
## SKA3.2 1.962031e-08 -0.4628294 0.005 0.055 7.181425e-04 2
## IFT43.1 1.991126e-08 0.4764670 0.024 0.090 7.287920e-04 2
## CREM.1 1.994115e-08 0.9535959 0.192 0.371 7.298860e-04 2
## SPAG5.2 1.998127e-08 -3.5592208 0.002 0.049 7.313543e-04 2
## AP003774.2 2.005927e-08 -0.8603954 0.008 0.060 7.342095e-04 2
## TBC1D7 2.011779e-08 0.4761418 0.039 0.115 7.363514e-04 2
## CASP7 2.053802e-08 0.5950955 0.045 0.124 7.517326e-04 2
## ZMYND11 2.058320e-08 0.9385046 0.126 0.256 7.533864e-04 2
## TSPAN14.1 2.062771e-08 0.9602206 0.177 0.344 7.550156e-04 2
## CYP51A1 2.088902e-08 0.4142112 0.045 0.123 7.645799e-04 2
## SELENOO 2.113323e-08 0.9676829 0.057 0.144 7.735186e-04 2
## SFXN3 2.140676e-08 0.3590297 0.026 0.092 7.835303e-04 2
## GLE1 2.175996e-08 0.7235239 0.051 0.134 7.964581e-04 2
## SPART.1 2.193133e-08 0.5115249 0.035 0.107 8.027307e-04 2
## HIF1AN 2.198201e-08 0.3498276 0.033 0.104 8.045855e-04 2
## SEPTIN8.1 2.201315e-08 -0.6993573 0.009 0.063 8.057253e-04 2
## CD2AP.2 2.260661e-08 0.9566111 0.132 0.267 8.274471e-04 2
## ACAD8 2.282823e-08 0.1127263 0.026 0.092 8.355589e-04 2
## GCH1.1 2.288694e-08 1.0962027 0.102 0.215 8.377079e-04 2
## ZBED5 2.289362e-08 0.6790121 0.053 0.136 8.379523e-04 2
## LASP1.1 2.294396e-08 0.9844246 0.164 0.320 8.397948e-04 2
## CCDC186.1 2.332448e-08 0.7356338 0.092 0.198 8.537227e-04 2
## ASB1 2.365520e-08 0.2947678 0.024 0.089 8.658275e-04 2
## BAG3.2 2.379417e-08 0.2670500 0.027 0.094 8.709143e-04 2
## GTF2I.1 2.383672e-08 0.7446979 0.185 0.353 8.724717e-04 2
## EIF2AK4 2.403820e-08 0.3045007 0.032 0.102 8.798462e-04 2
## POLRMT 2.408954e-08 0.3506772 0.026 0.092 8.817255e-04 2
## BAG4.1 2.429843e-08 0.4826067 0.045 0.123 8.893710e-04 2
## PYCR1.2 2.432320e-08 -0.2014329 0.005 0.054 8.902779e-04 2
## LRRC58.2 2.439954e-08 0.8676261 0.062 0.150 8.930721e-04 2
## VIM-AS1 2.453913e-08 0.6097065 0.065 0.155 8.981813e-04 2
## THUMPD3-AS1.1 2.490004e-08 0.8946481 0.081 0.182 9.113914e-04 2
## GLYR1.1 2.508889e-08 0.9592682 0.096 0.206 9.183035e-04 2
## CDK7 2.512225e-08 0.3345168 0.033 0.104 9.195246e-04 2
## DDX51 2.520785e-08 0.2891233 0.018 0.079 9.226578e-04 2
## AGPAT5 2.534264e-08 0.8885039 0.054 0.138 9.275913e-04 2
## WASHC2C.1 2.543741e-08 0.6374180 0.065 0.155 9.310601e-04 2
## NFKB2.1 2.550208e-08 0.9277380 0.122 0.248 9.334272e-04 2
## NSG1.1 2.566091e-08 -1.1984505 0.012 0.068 9.392407e-04 2
## MGST2.1 2.572935e-08 -1.5398345 0.006 0.057 9.417457e-04 2
## PRXL2B 2.580467e-08 -1.2795143 0.011 0.065 9.445026e-04 2
## MFN2 2.617694e-08 0.3861216 0.023 0.086 9.581284e-04 2
## CDS2.1 2.619453e-08 0.9104649 0.101 0.213 9.587722e-04 2
## RSU1.1 2.637660e-08 0.8402834 0.186 0.359 9.654363e-04 2
## FHIT.2 2.698291e-08 1.1770691 0.120 0.244 9.876283e-04 2
## PLK4.2 2.713203e-08 -2.3603382 0.005 0.054 9.930865e-04 2
## LNPK 2.733489e-08 0.5871195 0.048 0.128 1.000512e-03 2
## KLF12 2.752762e-08 2.0241023 0.377 0.398 1.007566e-03 2
## KCMF1.1 2.755818e-08 0.9266461 0.125 0.252 1.008684e-03 2
## CCDC69.1 2.755854e-08 1.0063140 0.122 0.247 1.008698e-03 2
## PPAT.1 2.766947e-08 0.6669716 0.038 0.111 1.012758e-03 2
## HNRNPA1P48 2.789405e-08 -1.2775379 0.008 0.059 1.020978e-03 2
## KLHDC7B 2.824464e-08 -1.6730724 0.006 0.057 1.033810e-03 2
## C2orf76 2.847190e-08 -0.3774594 0.008 0.059 1.042129e-03 2
## TOX.2 2.874706e-08 2.9750521 0.107 0.057 1.052200e-03 2
## TTC19 2.875216e-08 0.9580812 0.162 0.316 1.052387e-03 2
## PECR 2.893470e-08 0.5611369 0.048 0.128 1.059068e-03 2
## ZNF227 2.940096e-08 -0.8160427 0.009 0.062 1.076134e-03 2
## RELL2 2.949982e-08 0.5799795 0.039 0.114 1.079753e-03 2
## MIS12.1 2.994711e-08 0.4770529 0.044 0.120 1.096124e-03 2
## SLC35B3 3.001884e-08 0.8056030 0.036 0.109 1.098750e-03 2
## TMEM267 3.015270e-08 0.2158866 0.030 0.099 1.103649e-03 2
## BCL10 3.051537e-08 0.6124182 0.050 0.131 1.116923e-03 2
## INO80B 3.070862e-08 0.3162561 0.029 0.096 1.123997e-03 2
## NUP85.1 3.072030e-08 0.1017351 0.027 0.093 1.124425e-03 2
## CCDC51 3.091739e-08 0.1195803 0.011 0.065 1.131638e-03 2
## ABCB7 3.094648e-08 0.7478343 0.068 0.159 1.132703e-03 2
## PDCD4-AS1 3.138880e-08 -1.1732595 0.008 0.059 1.148893e-03 2
## S100B.2 3.143121e-08 -2.2768754 0.008 0.059 1.150445e-03 2
## RFT1.1 3.157289e-08 0.2933959 0.026 0.091 1.155631e-03 2
## PDE12.1 3.169643e-08 0.4520889 0.039 0.113 1.160153e-03 2
## CD58.1 3.203115e-08 0.8950508 0.123 0.249 1.172404e-03 2
## ADD1.2 3.301230e-08 0.7085897 0.235 0.441 1.208316e-03 2
## STK11.1 3.306354e-08 0.6330461 0.048 0.127 1.210192e-03 2
## CDCA3.2 3.308888e-08 -1.3632563 0.003 0.050 1.211119e-03 2
## IWS1.1 3.312663e-08 1.0323757 0.134 0.266 1.212501e-03 2
## CYTIP.2 3.323304e-08 0.5573824 0.334 0.631 1.216396e-03 2
## PCNX2 3.328224e-08 2.1782573 0.284 0.249 1.218196e-03 2
## B9D2 3.349067e-08 -0.1130205 0.017 0.075 1.225826e-03 2
## COX15 3.369703e-08 0.5936183 0.051 0.132 1.233379e-03 2
## PCBP1-AS1 3.374031e-08 0.9069666 0.066 0.157 1.234963e-03 2
## CAST.2 3.392070e-08 0.6330485 0.296 0.554 1.241566e-03 2
## HIF1A.1 3.407388e-08 0.7420248 0.179 0.344 1.247172e-03 2
## SLC43A1 3.469029e-08 0.3566178 0.024 0.088 1.269734e-03 2
## CNEP1R1 3.547942e-08 -0.7341104 0.012 0.067 1.298618e-03 2
## PRMT7.1 3.555609e-08 0.8030543 0.063 0.151 1.301424e-03 2
## ZNF263 3.615611e-08 0.1651792 0.020 0.080 1.323386e-03 2
## GNPNAT1.1 3.645160e-08 -0.2270722 0.015 0.072 1.334201e-03 2
## NCAPH2.2 3.698699e-08 0.5648059 0.026 0.090 1.353798e-03 2
## AC008892.1.1 3.706554e-08 -0.2239549 0.023 0.085 1.356673e-03 2
## LZTFL1.2 3.707782e-08 0.8120144 0.071 0.163 1.357122e-03 2
## HUS1 3.729158e-08 0.4233159 0.041 0.115 1.364946e-03 2
## TRNT1.1 3.751910e-08 0.8605130 0.071 0.163 1.373274e-03 2
## PIP4P2.1 3.790098e-08 -0.1211805 0.023 0.085 1.387252e-03 2
## PRKAG2 3.800446e-08 0.3874764 0.036 0.108 1.391039e-03 2
## GM2A.1 3.853997e-08 0.3974842 0.018 0.077 1.410640e-03 2
## NDFIP2.1 3.884429e-08 0.3036500 0.039 0.112 1.421779e-03 2
## EHD1.1 3.904883e-08 1.0171582 0.132 0.263 1.429265e-03 2
## PM20D2.1 3.970909e-08 -1.0299058 0.011 0.064 1.453432e-03 2
## TMEM186 3.987895e-08 -1.4892538 0.006 0.056 1.459649e-03 2
## CNOT10 4.008940e-08 0.9278724 0.078 0.175 1.467352e-03 2
## LRPPRC.2 4.010283e-08 0.6952644 0.132 0.263 1.467844e-03 2
## AGL 4.014225e-08 0.4944213 0.048 0.127 1.469287e-03 2
## TTC9C.1 4.038834e-08 0.1810582 0.023 0.085 1.478294e-03 2
## ALG13 4.060669e-08 0.8527310 0.116 0.236 1.486286e-03 2
## BORA.1 4.073376e-08 -2.0243246 0.003 0.050 1.490937e-03 2
## CIDEB.1 4.114315e-08 0.6551071 0.050 0.129 1.505922e-03 2
## WASHC2A 4.235437e-08 0.5460026 0.051 0.131 1.550255e-03 2
## NEU1.1 4.251748e-08 0.8889530 0.096 0.204 1.556225e-03 2
## GNB1L 4.267280e-08 0.1713679 0.018 0.077 1.561910e-03 2
## SMUG1.1 4.295248e-08 -0.5309187 0.014 0.069 1.572147e-03 2
## VKORC1L1.1 4.333546e-08 0.5649776 0.048 0.127 1.586165e-03 2
## ATPAF2 4.362794e-08 0.2071058 0.017 0.075 1.596870e-03 2
## KCTD17.1 4.377166e-08 0.4312475 0.021 0.082 1.602130e-03 2
## RCC1.2 4.380166e-08 0.9439698 0.036 0.107 1.603228e-03 2
## DHRS4L2.1 4.401083e-08 0.2277955 0.017 0.075 1.610884e-03 2
## PRPF3.1 4.416114e-08 0.8303932 0.078 0.175 1.616386e-03 2
## TNFRSF4.2 4.433455e-08 -1.2178470 0.011 0.064 1.622733e-03 2
## BLZF1.2 4.436699e-08 0.5146447 0.059 0.144 1.623921e-03 2
## CHIC2.1 4.439343e-08 0.9182733 0.110 0.225 1.624888e-03 2
## CORO7.2 4.457667e-08 0.7878761 0.233 0.437 1.631595e-03 2
## FDX2.1 4.458095e-08 -0.1633638 0.014 0.069 1.631752e-03 2
## SATB1.1 4.510611e-08 0.8354890 0.224 0.420 1.650974e-03 2
## USP11.2 4.528215e-08 0.7860431 0.065 0.153 1.657417e-03 2
## TFB1M 4.545088e-08 0.5313415 0.059 0.144 1.663593e-03 2
## MRM1.1 4.560817e-08 -1.4749151 0.005 0.052 1.669350e-03 2
## NAA16 4.562842e-08 0.8172580 0.071 0.162 1.670091e-03 2
## INAFM1.2 4.564201e-08 -0.6517109 0.005 0.052 1.670589e-03 2
## TSPAN4.1 4.582520e-08 -0.7052135 0.008 0.058 1.677294e-03 2
## INSIG2 4.593070e-08 0.4423246 0.033 0.102 1.681156e-03 2
## BCS1L.1 4.624300e-08 0.2165186 0.023 0.084 1.692586e-03 2
## MED13L.1 4.670107e-08 2.0913012 0.320 0.304 1.709353e-03 2
## GNGT2.2 4.677248e-08 -0.8144336 0.012 0.066 1.711966e-03 2
## NSMF 4.759347e-08 0.9500022 0.069 0.160 1.742016e-03 2
## SRRD 4.763110e-08 -0.3048604 0.020 0.079 1.743394e-03 2
## PTPN2.1 4.775676e-08 0.8193675 0.176 0.334 1.747993e-03 2
## HMGXB4.2 4.804595e-08 0.7542625 0.063 0.150 1.758578e-03 2
## CALU.1 4.825116e-08 0.9234697 0.077 0.172 1.766089e-03 2
## SH2D3A.1 4.885585e-08 0.5318307 0.033 0.102 1.788222e-03 2
## DELE1.1 4.928768e-08 0.6559639 0.090 0.193 1.804028e-03 2
## TRIM65 5.047641e-08 -0.4664185 0.020 0.079 1.847538e-03 2
## AC020915.2 5.048316e-08 -1.2673638 0.003 0.049 1.847785e-03 2
## PPP2R2D 5.075846e-08 0.8321619 0.077 0.172 1.857861e-03 2
## LMO4.1 5.187905e-08 0.8646152 0.060 0.145 1.898877e-03 2
## LIMK2.2 5.252871e-08 0.8252598 0.044 0.119 1.922656e-03 2
## DXO 5.345677e-08 0.5564773 0.027 0.092 1.956625e-03 2
## CSNK2A1 5.374455e-08 0.7769103 0.060 0.145 1.967158e-03 2
## UNC93B1.1 5.381992e-08 0.2647522 0.032 0.099 1.969917e-03 2
## CCNB2.2 5.399519e-08 -0.6829841 0.011 0.063 1.976332e-03 2
## THOC1.2 5.433099e-08 0.9261220 0.071 0.162 1.988623e-03 2
## BOD1 5.474266e-08 -0.2810051 0.018 0.076 2.003691e-03 2
## ITPK1.1 5.491955e-08 0.6996949 0.054 0.135 2.010166e-03 2
## GNA15.2 5.537005e-08 1.0205473 0.044 0.119 2.026655e-03 2
## THAP12 5.548233e-08 0.9302519 0.083 0.181 2.030764e-03 2
## NUP133.1 5.559309e-08 0.6185745 0.056 0.137 2.034818e-03 2
## ZNF48 5.575461e-08 -0.8374521 0.012 0.066 2.040730e-03 2
## ANAPC10 5.588855e-08 0.7427139 0.059 0.143 2.045633e-03 2
## MED17 5.615969e-08 0.3873694 0.063 0.149 2.055557e-03 2
## AL136456.1.2 5.652143e-08 3.0626504 0.141 0.086 2.068798e-03 2
## BAZ1A.1 5.663371e-08 0.8997180 0.200 0.377 2.072907e-03 2
## PDE6B.1 5.781728e-08 -1.1208064 0.009 0.060 2.116228e-03 2
## MOSMO.1 5.849675e-08 0.6825098 0.038 0.109 2.141098e-03 2
## FBXO22.1 5.851523e-08 0.7385646 0.057 0.140 2.141774e-03 2
## AL109840.2 5.864132e-08 4.4144362 0.021 0.004 2.146389e-03 2
## CAMK2D.1 5.931982e-08 1.9122133 0.368 0.384 2.171224e-03 2
## EGLN1 5.957317e-08 0.7198352 0.063 0.150 2.180497e-03 2
## SPTLC1 6.000012e-08 0.7341853 0.086 0.185 2.196124e-03 2
## C3orf18 6.011060e-08 -0.2343801 0.020 0.079 2.200168e-03 2
## UTP15.1 6.030984e-08 0.1498227 0.020 0.079 2.207461e-03 2
## ZBTB48 6.074670e-08 0.9043952 0.026 0.089 2.223451e-03 2
## MUTYH 6.118880e-08 0.9580571 0.042 0.116 2.239633e-03 2
## JMJD1C.1 6.122490e-08 1.7408758 0.421 0.488 2.240954e-03 2
## TMEM121 6.131555e-08 -0.8264715 0.009 0.060 2.244272e-03 2
## STK32C 6.177296e-08 0.2388117 0.021 0.081 2.261014e-03 2
## METTL3 6.204563e-08 0.5197598 0.047 0.123 2.270994e-03 2
## TTK.2 6.231798e-08 -0.8089084 0.003 0.049 2.280963e-03 2
## GCSAM 6.243235e-08 -2.8114890 0.002 0.046 2.285149e-03 2
## EDEM1 6.259193e-08 0.7065230 0.068 0.157 2.290990e-03 2
## CUL5.1 6.286217e-08 0.9308652 0.125 0.250 2.300881e-03 2
## ORC4 6.312447e-08 0.7446454 0.063 0.150 2.310482e-03 2
## SLC25A12.1 6.350022e-08 0.9050731 0.089 0.190 2.324235e-03 2
## CYHR1 6.513845e-08 0.7283666 0.038 0.109 2.384198e-03 2
## ECE1.2 6.597763e-08 0.7185555 0.072 0.163 2.414913e-03 2
## TTC9 6.656873e-08 0.9620122 0.068 0.157 2.436549e-03 2
## NUP155.2 6.801517e-08 0.7186712 0.036 0.106 2.489491e-03 2
## RAF1.1 6.823569e-08 0.9903434 0.089 0.190 2.497563e-03 2
## NVL 6.928625e-08 0.7480214 0.078 0.172 2.536015e-03 2
## ID3.2 6.953474e-08 -5.6277696 0.000 0.042 2.545111e-03 2
## FRAT2 6.957429e-08 -0.6256190 0.014 0.068 2.546558e-03 2
## DOLPP1 6.993922e-08 -0.6484402 0.006 0.054 2.559915e-03 2
## CALCOCO1.2 7.026834e-08 0.7487615 0.065 0.152 2.571962e-03 2
## NKRF 7.029305e-08 -0.2764005 0.015 0.071 2.572866e-03 2
## CCNH.1 7.077023e-08 0.8886299 0.179 0.338 2.590332e-03 2
## MANBAL 7.086095e-08 -0.4657571 0.017 0.073 2.593653e-03 2
## WRAP53.1 7.104455e-08 0.3968510 0.014 0.068 2.600373e-03 2
## DENND6A 7.162894e-08 0.6738558 0.095 0.199 2.621762e-03 2
## CSRNP1.1 7.185841e-08 0.5954307 0.057 0.140 2.630161e-03 2
## GTSF1 7.322951e-08 -2.0239833 0.005 0.051 2.680347e-03 2
## ACAD9 7.327434e-08 0.6040367 0.032 0.098 2.681987e-03 2
## PAFAH1B2 7.385815e-08 0.5766048 0.044 0.118 2.703356e-03 2
## KAT5 7.406331e-08 0.3763592 0.041 0.113 2.710865e-03 2
## ATP6V1C1 7.412948e-08 0.4009406 0.051 0.130 2.713287e-03 2
## PTBP3.1 7.414441e-08 0.7015163 0.263 0.493 2.713834e-03 2
## KIF4A.2 7.434827e-08 -0.4636483 0.005 0.051 2.721296e-03 2
## PHLDA2.1 7.526282e-08 -0.7663253 0.006 0.054 2.754770e-03 2
## PVT1 7.556492e-08 1.8181745 0.402 0.441 2.765827e-03 2
## CDK11B.1 7.561447e-08 0.8128352 0.077 0.170 2.767641e-03 2
## STK19 7.618823e-08 0.6831304 0.029 0.093 2.788641e-03 2
## VOPP1.1 7.733852e-08 0.9336092 0.203 0.381 2.830744e-03 2
## NUP160.2 7.790073e-08 0.8147674 0.078 0.172 2.851322e-03 2
## STT3B 7.892824e-08 0.8144892 0.224 0.419 2.888931e-03 2
## ASPHD2 7.968392e-08 -0.9913416 0.008 0.057 2.916591e-03 2
## ARHGAP27 8.172073e-08 0.8899934 0.042 0.115 2.991142e-03 2
## ZNF274 8.211761e-08 0.5494161 0.030 0.095 3.005669e-03 2
## GNPTAB.2 8.235636e-08 0.8212301 0.192 0.361 3.014407e-03 2
## RBL1.2 8.304634e-08 0.7634414 0.089 0.188 3.039662e-03 2
## PRPF18 8.342152e-08 0.6335694 0.053 0.132 3.053395e-03 2
## MON1A 8.382814e-08 -0.6161723 0.008 0.056 3.068278e-03 2
## ABHD12.2 8.474966e-08 0.8836951 0.054 0.134 3.102007e-03 2
## FBXL12 8.506360e-08 0.5899116 0.027 0.090 3.113498e-03 2
## ZNF408 8.517783e-08 -0.6661202 0.011 0.062 3.117679e-03 2
## ZNF714.2 8.536866e-08 0.1421949 0.024 0.085 3.124664e-03 2
## ARHGAP17.1 8.551591e-08 0.7463031 0.081 0.177 3.130053e-03 2
## ATP6V1A 8.554958e-08 0.8244576 0.056 0.136 3.131286e-03 2
## CCAR2 8.613164e-08 0.6772554 0.083 0.179 3.152590e-03 2
## HIST2H2AC 8.669295e-08 1.0713863 0.045 0.120 3.173136e-03 2
## SLAMF6.2 8.707977e-08 0.7246613 0.071 0.160 3.187294e-03 2
## SHLD1.1 8.861418e-08 0.9526587 0.033 0.100 3.243456e-03 2
## FBXO44.1 8.886389e-08 0.5660396 0.032 0.098 3.252596e-03 2
## NFX1 8.923516e-08 0.7995035 0.107 0.217 3.266185e-03 2
## DVL2.1 9.026440e-08 0.1396825 0.015 0.070 3.303857e-03 2
## C16orf58 9.073327e-08 0.2843052 0.023 0.083 3.321019e-03 2
## PTEN 9.099348e-08 0.8297921 0.211 0.391 3.330543e-03 2
## SNAPC1.1 9.140483e-08 0.5246385 0.026 0.088 3.345600e-03 2
## CLU.1 9.181032e-08 -0.3813299 0.008 0.056 3.360441e-03 2
## ZNF296.1 9.198717e-08 -0.5991599 0.008 0.056 3.366915e-03 2
## RSPRY1 9.351586e-08 0.3388682 0.051 0.129 3.422867e-03 2
## ERCC2.1 9.379614e-08 0.3325603 0.015 0.070 3.433126e-03 2
## MORC3 9.427100e-08 0.6940035 0.117 0.235 3.450507e-03 2
## ELMOD2.1 9.474056e-08 -0.4388242 0.017 0.072 3.467694e-03 2
## BAP1 9.551957e-08 0.1020369 0.026 0.087 3.496207e-03 2
## TTF2.2 9.552063e-08 0.9296998 0.074 0.164 3.496246e-03 2
## ANKLE2 9.599252e-08 0.8396075 0.110 0.221 3.513518e-03 2
## SWSAP1 9.615276e-08 -0.4436940 0.009 0.059 3.519383e-03 2
## SPINDOC.1 9.669199e-08 0.5608212 0.018 0.075 3.539120e-03 2
## NCKAP1L.2 9.706219e-08 0.7551133 0.200 0.376 3.552670e-03 2
## CEP128.2 9.714577e-08 0.5256800 0.066 0.152 3.555729e-03 2
## CDADC1 9.749223e-08 0.4437074 0.023 0.082 3.568411e-03 2
## DNAJC3.1 9.794447e-08 0.8401194 0.177 0.335 3.584963e-03 2
## TOP1MT.1 9.889751e-08 -5.3377494 0.000 0.041 3.619847e-03 2
## TOM1 9.900649e-08 0.6395512 0.062 0.145 3.623835e-03 2
## RSBN1L 9.919402e-08 1.0208990 0.186 0.352 3.630700e-03 2
## EIF2B4 9.958369e-08 0.5868567 0.041 0.112 3.644962e-03 2
## UBL3.1 1.005827e-07 0.6614634 0.090 0.191 3.681527e-03 2
## ZNF574 1.009692e-07 0.2671530 0.023 0.082 3.695675e-03 2
## ZBED1 1.013090e-07 0.6703306 0.038 0.107 3.708112e-03 2
## PTP4A3 1.013139e-07 -0.2106797 0.017 0.072 3.708290e-03 2
## CNP.2 1.015296e-07 0.6518248 0.030 0.095 3.716187e-03 2
## PALM2-AKAP2.2 1.016514e-07 0.7677317 0.074 0.164 3.720644e-03 2
## BUB1B.2 1.017863e-07 -0.2309022 0.015 0.069 3.725582e-03 2
## WDR62.2 1.019992e-07 -1.0616785 0.006 0.053 3.733376e-03 2
## SPSB2 1.020415e-07 -0.7030314 0.009 0.059 3.734921e-03 2
## AFTPH 1.027099e-07 0.7897877 0.105 0.215 3.759386e-03 2
## C1orf216.1 1.035395e-07 -0.6982530 0.008 0.056 3.789752e-03 2
## RNF41 1.038005e-07 0.7481505 0.042 0.114 3.799307e-03 2
## GSR.1 1.048480e-07 0.8678680 0.086 0.183 3.837646e-03 2
## HSPA14.1.1 1.051218e-07 0.4988712 0.020 0.077 3.847668e-03 2
## FCF1.1 1.053340e-07 0.4749329 0.033 0.099 3.855434e-03 2
## BRD8 1.055025e-07 0.6137190 0.060 0.142 3.861601e-03 2
## CCDC102A 1.062866e-07 0.1997884 0.018 0.075 3.890301e-03 2
## HARS2 1.070860e-07 0.2073896 0.030 0.094 3.919562e-03 2
## STK17A.1 1.081437e-07 0.8260219 0.265 0.490 3.958274e-03 2
## USP37.2 1.085724e-07 1.0253692 0.062 0.145 3.973967e-03 2
## SUV39H2.2 1.086059e-07 -1.5807881 0.005 0.050 3.975193e-03 2
## MFSD11 1.086991e-07 1.1126948 0.090 0.191 3.978604e-03 2
## FUBP3.1 1.094403e-07 0.8745528 0.051 0.128 4.005735e-03 2
## SHCBP1.2 1.094492e-07 0.2138462 0.014 0.067 4.006060e-03 2
## SEC24C 1.099189e-07 1.0675692 0.093 0.196 4.023251e-03 2
## CRYZL1 1.099450e-07 0.7298751 0.047 0.121 4.024206e-03 2
## ATXN10 1.110694e-07 0.9584201 0.146 0.281 4.065363e-03 2
## TERF2 1.111089e-07 0.8332221 0.050 0.126 4.066809e-03 2
## PHF10.1 1.117995e-07 0.6013838 0.054 0.133 4.092084e-03 2
## TMEM43.1 1.129048e-07 0.7725027 0.078 0.171 4.132543e-03 2
## SPRYD7.1 1.135958e-07 -1.3812415 0.006 0.053 4.157835e-03 2
## CTU2 1.141699e-07 0.4608406 0.014 0.067 4.178847e-03 2
## MAPK7 1.152848e-07 -0.5627262 0.011 0.061 4.219653e-03 2
## APLP2.1 1.155813e-07 0.8359067 0.051 0.128 4.230508e-03 2
## HVCN1.2 1.156331e-07 -0.3114594 0.017 0.072 4.232402e-03 2
## KNSTRN.2 1.157897e-07 -2.1246041 0.003 0.047 4.238134e-03 2
## PXK.1 1.158791e-07 0.8663082 0.060 0.143 4.241407e-03 2
## FAAH2.2 1.170047e-07 0.1908663 0.026 0.087 4.282606e-03 2
## ARGLU1 1.176838e-07 1.3534912 0.493 0.632 4.307462e-03 2
## RCC2.2 1.178947e-07 0.5554272 0.060 0.142 4.315183e-03 2
## WDHD1.2 1.189305e-07 0.6911171 0.038 0.107 4.353095e-03 2
## PRORP 1.194620e-07 1.0509593 0.137 0.266 4.372549e-03 2
## TTF1.1 1.195013e-07 0.5482653 0.057 0.137 4.373987e-03 2
## CERS4.1 1.203740e-07 1.0339585 0.059 0.140 4.405928e-03 2
## CEP135.2 1.216935e-07 0.3927574 0.038 0.106 4.454227e-03 2
## DONSON.2 1.228857e-07 0.4785161 0.026 0.087 4.497863e-03 2
## LRRC45.2 1.232162e-07 0.8867066 0.035 0.102 4.509959e-03 2
## AKR1C3.1 1.242475e-07 -0.8919582 0.009 0.058 4.547705e-03 2
## HNRNPH1 1.246720e-07 0.7538855 0.223 0.412 4.563244e-03 2
## MAP7D3 1.249441e-07 0.5149714 0.036 0.104 4.573204e-03 2
## KIF18B.2 1.261544e-07 -2.2988120 0.002 0.044 4.617502e-03 2
## NT5E.1 1.266574e-07 -0.3055498 0.021 0.079 4.635915e-03 2
## TASOR.1 1.267093e-07 0.9255272 0.150 0.289 4.637814e-03 2
## SRR.2 1.270998e-07 -0.3232486 0.008 0.055 4.652108e-03 2
## SCO1 1.292485e-07 0.7822451 0.038 0.106 4.730754e-03 2
## AC005842.1.2 1.293038e-07 0.5473965 0.011 0.061 4.732777e-03 2
## TAF2.1 1.293532e-07 0.7782794 0.075 0.166 4.734587e-03 2
## PCYOX1L.1 1.299784e-07 0.8860740 0.056 0.135 4.757471e-03 2
## MEMO1 1.304634e-07 -0.3069089 0.017 0.071 4.775222e-03 2
## MED20 1.307257e-07 -0.1164327 0.014 0.066 4.784820e-03 2
## ZNF862 1.311775e-07 0.8093731 0.047 0.120 4.801360e-03 2
## CCHCR1.1 1.344684e-07 -0.2545268 0.009 0.058 4.921813e-03 2
## HERPUD2.2 1.351841e-07 0.9021308 0.125 0.245 4.948008e-03 2
## XKR8.1 1.360515e-07 -1.2068679 0.008 0.055 4.979758e-03 2
## PNMA1 1.365847e-07 0.2169728 0.029 0.091 4.999272e-03 2
## POC5.1 1.370372e-07 0.6663776 0.051 0.128 5.015835e-03 2
## FCHO1 1.377763e-07 0.9774806 0.053 0.130 5.042887e-03 2
## MICU2.1 1.378600e-07 0.8856174 0.173 0.326 5.045951e-03 2
## DHX8 1.386142e-07 0.6181556 0.051 0.128 5.073556e-03 2
## DNAAF5.1 1.386768e-07 0.5450780 0.014 0.066 5.075847e-03 2
## MX1.2 1.393193e-07 0.8122420 0.050 0.125 5.099367e-03 2
## KCNN4 1.401530e-07 0.1004164 0.027 0.089 5.129880e-03 2
## BOLA1 1.409063e-07 0.9908791 0.039 0.109 5.157453e-03 2
## WDFY1.1 1.421221e-07 0.7913749 0.089 0.187 5.201953e-03 2
## PRR3 1.431152e-07 0.8044663 0.045 0.118 5.238301e-03 2
## SGTB 1.433133e-07 0.5305891 0.050 0.125 5.245553e-03 2
## ARHGAP11A.2 1.458485e-07 0.3262643 0.027 0.089 5.338348e-03 2
## CRCP 1.468296e-07 0.9648156 0.080 0.172 5.374256e-03 2
## CCNE1.2 1.505341e-07 -0.1050819 0.006 0.052 5.509849e-03 2
## SERTAD3.1 1.508400e-07 -0.1105663 0.017 0.071 5.521046e-03 2
## QSOX1 1.518774e-07 0.2987861 0.033 0.098 5.559018e-03 2
## DOCK2.1 1.524361e-07 1.7862822 0.417 0.482 5.579466e-03 2
## MMP25-AS1.1 1.525461e-07 0.8407408 0.120 0.236 5.583491e-03 2
## TNFRSF10A 1.529454e-07 0.6456606 0.051 0.127 5.598109e-03 2
## TRAPPC6B.1 1.541352e-07 0.7160678 0.056 0.134 5.641656e-03 2
## STX2 1.548478e-07 0.4725616 0.042 0.113 5.667741e-03 2
## VRK2.1 1.555913e-07 0.7456372 0.060 0.141 5.694952e-03 2
## ZNF195 1.566331e-07 0.6788102 0.053 0.129 5.733084e-03 2
## IL9R.1 1.568004e-07 0.4059539 0.089 0.184 5.739207e-03 2
## TNPO1.1 1.571964e-07 0.9293596 0.101 0.206 5.753703e-03 2
## CLK2 1.578031e-07 0.7400545 0.053 0.130 5.775910e-03 2
## C3orf14.1 1.609115e-07 -0.6889679 0.006 0.052 5.889684e-03 2
## POLE2.2 1.635530e-07 -0.3129838 0.009 0.057 5.986368e-03 2
## CETN2 1.658116e-07 -1.7054194 0.003 0.046 6.069037e-03 2
## HSD17B7.1 1.661364e-07 0.2796156 0.029 0.091 6.080925e-03 2
## HELLS.2 1.663669e-07 0.9033487 0.089 0.185 6.089360e-03 2
## MHENCR.1 1.686272e-07 0.2269799 0.026 0.086 6.172091e-03 2
## DPH7 1.696502e-07 0.8770111 0.047 0.120 6.209535e-03 2
## SLC25A17.1 1.714962e-07 0.9941448 0.042 0.113 6.277103e-03 2
## AC008105.3.1 1.716774e-07 0.9667616 0.116 0.229 6.283737e-03 2
## RNF6.1 1.719561e-07 0.6320686 0.065 0.148 6.293935e-03 2
## TMEM68 1.726452e-07 0.3542339 0.029 0.091 6.319161e-03 2
## ASB6 1.737455e-07 0.6253340 0.039 0.107 6.359432e-03 2
## ATP2B1-AS1 1.739090e-07 0.8363063 0.036 0.103 6.365417e-03 2
## INPP4A.2 1.749695e-07 1.9015504 0.374 0.398 6.404232e-03 2
## TACC1.1 1.756179e-07 0.9299388 0.188 0.353 6.427967e-03 2
## PTPN13.2 1.774075e-07 3.1788465 0.039 0.013 6.493469e-03 2
## EED.1 1.801826e-07 0.7871489 0.083 0.176 6.595044e-03 2
## RBM48 1.814942e-07 0.4179590 0.036 0.103 6.643050e-03 2
## ATG4C 1.816983e-07 0.7514667 0.024 0.083 6.650521e-03 2
## SSX2IP 1.826648e-07 -0.2274627 0.009 0.057 6.685897e-03 2
## MCOLN1 1.834548e-07 -1.3718027 0.005 0.049 6.714813e-03 2
## AKAP7 1.840050e-07 0.7047085 0.057 0.136 6.734949e-03 2
## ASL 1.843081e-07 0.9007465 0.053 0.129 6.746045e-03 2
## NCBP1 1.855279e-07 0.9022502 0.081 0.174 6.790690e-03 2
## NUDT9 1.859125e-07 0.2185205 0.021 0.078 6.804770e-03 2
## ROGDI 1.883397e-07 0.3623229 0.018 0.073 6.893609e-03 2
## POU2F2 1.892679e-07 0.9144317 0.092 0.191 6.927582e-03 2
## ZNF138.1 1.921001e-07 0.5376525 0.035 0.100 7.031248e-03 2
## ZBTB20.1 1.930496e-07 1.9476357 0.389 0.432 7.066000e-03 2
## PDP1 1.969285e-07 0.3629423 0.027 0.088 7.207977e-03 2
## FANCE.2 1.970292e-07 -0.8116468 0.005 0.048 7.211663e-03 2
## ELOA-AS1 1.988356e-07 0.3919267 0.020 0.075 7.277782e-03 2
## CCM2.1 1.988609e-07 0.9469854 0.239 0.443 7.278706e-03 2
## E2F2.2 2.006688e-07 -0.6349670 0.011 0.059 7.344880e-03 2
## GCC2.1 2.011746e-07 0.9290080 0.232 0.427 7.363392e-03 2
## SOAT1.1 2.021226e-07 0.6811274 0.080 0.172 7.398090e-03 2
## MAFG.2 2.025793e-07 0.4265325 0.024 0.083 7.414808e-03 2
## ITGA6.1 2.031961e-07 0.3882804 0.048 0.121 7.437384e-03 2
## E2F8.2 2.042805e-07 -1.2334405 0.003 0.045 7.477073e-03 2
## JUND.1 2.045181e-07 0.4248518 0.433 0.787 7.485771e-03 2
## TMPO-AS1.2 2.048234e-07 -0.8389563 0.003 0.045 7.496944e-03 2
## CES2 2.054584e-07 0.2390012 0.029 0.090 7.520189e-03 2
## TRAPPC13.1 2.055716e-07 0.1442108 0.024 0.083 7.524330e-03 2
## ZNF131 2.068615e-07 0.9729657 0.161 0.304 7.571544e-03 2
## TTLL12.2 2.080488e-07 0.2126221 0.024 0.083 7.615003e-03 2
## BCKDHB.1 2.084177e-07 0.8004301 0.063 0.145 7.628503e-03 2
## CACNA2D4 2.084784e-07 -0.9780692 0.006 0.051 7.630725e-03 2
## CYB561.1 2.091600e-07 -0.3375667 0.014 0.065 7.655673e-03 2
## ANKZF1 2.095298e-07 0.5724898 0.050 0.124 7.669210e-03 2
## TIMM21.1 2.099523e-07 0.6383551 0.023 0.080 7.684674e-03 2
## E4F1.1 2.106544e-07 0.9887487 0.035 0.100 7.710372e-03 2
## GPR183.2 2.120630e-07 0.7758534 0.098 0.199 7.761931e-03 2
## CMPK2.2 2.134444e-07 -0.4285986 0.009 0.057 7.812493e-03 2
## SLC15A4 2.137298e-07 0.2817409 0.033 0.097 7.822940e-03 2
## ZNF626 2.138448e-07 -1.0182061 0.011 0.059 7.827148e-03 2
## PCNT.1 2.143369e-07 0.9077786 0.107 0.213 7.845158e-03 2
## GTPBP10 2.168903e-07 0.7348770 0.069 0.154 7.938619e-03 2
## SIRT2 2.226071e-07 1.0893340 0.125 0.242 8.147865e-03 2
## TCHP 2.226959e-07 1.2844783 0.104 0.210 8.151115e-03 2
## PSMC3IP.2 2.270017e-07 0.2240715 0.005 0.048 8.308715e-03 2
## NUDT4.1 2.273592e-07 0.8726347 0.092 0.189 8.321800e-03 2
## CREG1.2 2.299914e-07 -0.8069793 0.009 0.056 8.418146e-03 2
## HIST1H3F.2 2.301172e-07 -0.4837839 0.017 0.069 8.422751e-03 2
## NGLY1 2.308226e-07 1.0706288 0.143 0.273 8.448569e-03 2
## AANAT 2.310593e-07 5.2931087 0.012 0.001 8.457231e-03 2
## LARP4.1 2.315989e-07 1.0654474 0.102 0.206 8.476983e-03 2
## GNS 2.324736e-07 0.5642594 0.041 0.109 8.508998e-03 2
## PANK4 2.336253e-07 0.1814164 0.026 0.085 8.551152e-03 2
## EIF5A2 2.337711e-07 -2.6857426 0.003 0.045 8.556488e-03 2
## NFRKB 2.345237e-07 0.6805296 0.059 0.138 8.584037e-03 2
## FUZ 2.352316e-07 -0.6049364 0.011 0.059 8.609945e-03 2
## ACTN1.2 2.354602e-07 -1.3596894 0.003 0.045 8.618313e-03 2
## SCD.2 2.355097e-07 -0.2337655 0.012 0.062 8.620126e-03 2
## LYRM7 2.363704e-07 0.4658821 0.033 0.097 8.651629e-03 2
## TRAV8-2.1 2.381846e-07 -0.1520962 0.027 0.087 8.718033e-03 2
## TRG-AS1.2 2.386951e-07 0.7728276 0.117 0.230 8.736717e-03 2
## DDX28 2.397347e-07 0.1649240 0.012 0.062 8.774770e-03 2
## PLPP1.2 2.399024e-07 0.7735383 0.093 0.191 8.780908e-03 2
## ZFP36L2.1 2.404033e-07 0.6303125 0.392 0.714 8.799241e-03 2
## CD3EAP.1 2.411563e-07 -1.4699797 0.006 0.051 8.826803e-03 2
## SLC38A1 2.424488e-07 1.4387198 0.490 0.650 8.874111e-03 2
## XYLT1.1 2.426233e-07 2.2333295 0.287 0.259 8.880498e-03 2
## FASTKD5 2.444405e-07 -0.8253751 0.009 0.056 8.947010e-03 2
## UCK2.2 2.450119e-07 0.7791525 0.032 0.095 8.967924e-03 2
## KCTD5 2.450269e-07 0.9476799 0.039 0.106 8.968476e-03 2
## MACO1 2.467878e-07 0.9309757 0.087 0.183 9.032927e-03 2
## RAD18.2 2.502004e-07 0.8143943 0.033 0.097 9.157836e-03 2
## FBXW2 2.506147e-07 0.8330791 0.084 0.178 9.173001e-03 2
## SMIM14 2.547664e-07 0.4675822 0.041 0.109 9.324960e-03 2
## ZNF12 2.550084e-07 0.6301358 0.035 0.099 9.333816e-03 2
## WDYHV1 2.558948e-07 0.3484624 0.020 0.075 9.366261e-03 2
## SRP14-AS1 2.566451e-07 -0.1813183 0.014 0.064 9.393724e-03 2
## PIP4K2A.1 2.568047e-07 1.5772147 0.441 0.537 9.399565e-03 2
## RORA 2.576640e-07 1.8805208 0.383 0.413 9.431018e-03 2
## ZNF641.1 2.608141e-07 -0.2075458 0.012 0.061 9.546319e-03 2
## TMEM268 2.624175e-07 0.2344124 0.026 0.084 9.605005e-03 2
## MON1B 2.627741e-07 0.8807472 0.066 0.149 9.618057e-03 2
## RPUSD2 2.632351e-07 -0.2376739 0.018 0.072 9.634931e-03 2
## PRIM2.2 2.636165e-07 1.0986527 0.086 0.180 9.648891e-03 2
## EXOSC2.2 2.677019e-07 0.5963122 0.035 0.099 9.798424e-03 2
## DERL3.1 2.697420e-07 -1.1287467 0.003 0.045 9.873097e-03 2
## AP5B1 2.724032e-07 -3.2310754 0.002 0.042 9.970503e-03 2
## GFPT1.1 2.795429e-07 0.7675135 0.090 0.187 1.023183e-02 2
## MIEF1 2.797880e-07 -0.5155605 0.021 0.076 1.024080e-02 2
## AGMAT 2.798222e-07 -1.2655121 0.006 0.050 1.024205e-02 2
## TAF1 2.816720e-07 0.9216114 0.081 0.172 1.030976e-02 2
## MED18 2.821089e-07 -0.2131639 0.023 0.079 1.032575e-02 2
## WDR43.1 2.843358e-07 0.8404334 0.110 0.218 1.040726e-02 2
## DNAJA1 2.856740e-07 0.8640259 0.307 0.571 1.045624e-02 2
## SCYL3 2.866113e-07 0.5113533 0.045 0.115 1.049055e-02 2
## ZNF224 2.892932e-07 0.2379910 0.032 0.093 1.058871e-02 2
## MAP2K7 2.893050e-07 0.6218948 0.048 0.120 1.058914e-02 2
## RECQL4.2 2.900016e-07 -0.1734732 0.008 0.053 1.061464e-02 2
## KIF3A 2.948003e-07 0.7655423 0.065 0.146 1.079028e-02 2
## NECAP1 2.955464e-07 0.6282710 0.068 0.151 1.081759e-02 2
## VAT1 2.960398e-07 -0.1363817 0.017 0.069 1.083565e-02 2
## SCYL2 2.972103e-07 0.6786391 0.071 0.156 1.087849e-02 2
## TOP2B 2.985036e-07 0.9286453 0.195 0.363 1.092583e-02 2
## POP1.1 3.001333e-07 0.3258685 0.026 0.084 1.098548e-02 2
## MBD6 3.001607e-07 0.6143579 0.032 0.094 1.098648e-02 2
## B3GNT8.1 3.005523e-07 -1.1848147 0.006 0.050 1.100081e-02 2
## AP001011.1 3.021560e-07 0.6881251 0.066 0.148 1.105951e-02 2
## TRAV16 3.038404e-07 -0.7325397 0.009 0.055 1.112117e-02 2
## FBXL5 3.058405e-07 1.0501481 0.105 0.210 1.119437e-02 2
## CCDC66 3.062985e-07 0.9672017 0.098 0.198 1.121114e-02 2
## ABHD13 3.074653e-07 0.6535347 0.047 0.118 1.125385e-02 2
## DSTYK 3.114064e-07 0.1430015 0.024 0.081 1.139810e-02 2
## TRIP6.2 3.133294e-07 -1.6306177 0.008 0.053 1.146848e-02 2
## DNAJC1.2 3.162369e-07 0.9791860 0.180 0.334 1.157490e-02 2
## TAF8 3.171442e-07 0.8581713 0.047 0.117 1.160811e-02 2
## DSCC1.2 3.173968e-07 0.6196624 0.008 0.053 1.161736e-02 2
## PRKAR2A 3.184056e-07 0.9301681 0.093 0.191 1.165428e-02 2
## PJA1 3.184974e-07 -0.2370891 0.020 0.074 1.165764e-02 2
## C12orf73 3.193787e-07 0.5716294 0.021 0.076 1.168990e-02 2
## AGK 3.201691e-07 0.7913191 0.060 0.139 1.171883e-02 2
## TRGC2.2 3.228030e-07 -0.2895694 0.014 0.063 1.181523e-02 2
## OFD1.1 3.234032e-07 1.0900590 0.128 0.247 1.183720e-02 2
## EEF1AKMT1 3.275045e-07 0.5873869 0.024 0.081 1.198732e-02 2
## FIGNL1.2 3.279019e-07 0.8936032 0.023 0.079 1.200187e-02 2
## AC010609.1.1 3.288786e-07 0.5895577 0.036 0.101 1.203762e-02 2
## HDAC10 3.288952e-07 0.6940563 0.038 0.103 1.203822e-02 2
## PITPNM1 3.324314e-07 0.8924799 0.045 0.115 1.216765e-02 2
## PIDD1.2 3.336110e-07 0.6720783 0.027 0.086 1.221083e-02 2
## SLC37A4.2 3.373027e-07 -0.6420067 0.008 0.053 1.234595e-02 2
## NECAB3.2 3.386275e-07 0.1247795 0.021 0.076 1.239444e-02 2
## RPUSD4 3.405759e-07 0.6933448 0.027 0.086 1.246576e-02 2
## CRKL 3.418029e-07 0.6756133 0.059 0.136 1.251067e-02 2
## FKBP5.2 3.473712e-07 0.9626759 0.182 0.338 1.271448e-02 2
## AC083798.2 3.477791e-07 -0.5044292 0.009 0.055 1.272941e-02 2
## MR1 3.493737e-07 0.9382189 0.065 0.145 1.278778e-02 2
## FBXO4.2 3.566026e-07 0.4684398 0.030 0.091 1.305237e-02 2
## GGA2 3.598554e-07 0.9327046 0.096 0.195 1.317143e-02 2
## SETD3 3.658287e-07 0.9646218 0.081 0.172 1.339006e-02 2
## ZNF367.2 3.662360e-07 0.8943312 0.027 0.086 1.340497e-02 2
## PORCN.1 3.671418e-07 0.2787812 0.029 0.088 1.343812e-02 2
## SLC35F6 3.688388e-07 -0.4527264 0.011 0.058 1.350024e-02 2
## YJU2 3.691961e-07 0.6163062 0.050 0.122 1.351332e-02 2
## TMEM192 3.713788e-07 0.7974966 0.026 0.083 1.359321e-02 2
## TTLL1 3.739683e-07 0.2394565 0.011 0.058 1.368799e-02 2
## AP3M2.2 3.746670e-07 0.9025271 0.066 0.148 1.371356e-02 2
## VPS13B.1 3.765045e-07 1.8925901 0.380 0.414 1.378082e-02 2
## NUP214 3.776207e-07 0.9967981 0.114 0.223 1.382167e-02 2
## CDC37L1 3.776903e-07 0.7421415 0.044 0.112 1.382422e-02 2
## AC008875.3 3.778953e-07 1.1520925 0.072 0.157 1.383172e-02 2
## SAYSD1 3.789698e-07 0.4999761 0.023 0.078 1.387105e-02 2
## USF3 3.793743e-07 0.8712106 0.060 0.138 1.388586e-02 2
## CXorf38 3.809272e-07 0.9867737 0.053 0.127 1.394270e-02 2
## MTO1 3.814139e-07 0.8685029 0.065 0.146 1.396051e-02 2
## METTL1.2 3.845967e-07 -1.6913482 0.005 0.047 1.407701e-02 2
## ENG 3.873087e-07 0.4885695 0.021 0.076 1.417627e-02 2
## ACP2 3.891413e-07 -0.2115427 0.024 0.081 1.424335e-02 2
## MBD1 3.908296e-07 0.9602981 0.083 0.173 1.430515e-02 2
## SP2 3.954198e-07 0.5191574 0.041 0.107 1.447316e-02 2
## CLCN3 3.965315e-07 0.9643179 0.110 0.216 1.451385e-02 2
## SLCO4A1.1 3.966331e-07 0.4755317 0.021 0.076 1.451757e-02 2
## SNX27 3.976934e-07 0.5103054 0.042 0.110 1.455637e-02 2
## GPR18.2 3.982522e-07 0.7253177 0.030 0.091 1.457683e-02 2
## RIF1.2 4.002926e-07 0.9773604 0.122 0.236 1.465151e-02 2
## CD9.2 4.033626e-07 -5.8267992 0.000 0.037 1.476388e-02 2
## PFAS.1 4.040286e-07 0.4376687 0.014 0.063 1.478825e-02 2
## RETREG1.1 4.058774e-07 0.9161139 0.099 0.199 1.485592e-02 2
## ZNF84.1 4.081046e-07 0.6368397 0.036 0.100 1.493745e-02 2
## HSD17B12.2 4.104037e-07 0.8923038 0.030 0.091 1.502160e-02 2
## NPTN.1 4.111358e-07 0.8523203 0.081 0.171 1.504839e-02 2
## COG1 4.147907e-07 0.4508269 0.027 0.086 1.518217e-02 2
## CUL3 4.182459e-07 0.9193796 0.215 0.396 1.530864e-02 2
## GPBP1 4.257234e-07 0.8524031 0.218 0.399 1.558233e-02 2
## MMS22L.2 4.264566e-07 0.6084064 0.053 0.126 1.560916e-02 2
## CYB5R4.1 4.274751e-07 0.9352134 0.077 0.164 1.564644e-02 2
## ZC3H6 4.279646e-07 0.6794096 0.048 0.119 1.566436e-02 2
## PPP2R5A 4.283384e-07 0.8251727 0.168 0.312 1.567804e-02 2
## EZH1.1 4.320717e-07 0.8174322 0.056 0.131 1.581469e-02 2
## WDR73 4.345148e-07 0.3690304 0.026 0.083 1.590411e-02 2
## ERCC4 4.362681e-07 0.2428176 0.023 0.078 1.596829e-02 2
## ZNF668.2 4.385342e-07 0.5704211 0.024 0.080 1.605123e-02 2
## IL23A.1 4.463425e-07 0.8624048 0.026 0.083 1.633703e-02 2
## TMEM87A 4.496791e-07 0.9989698 0.141 0.268 1.645915e-02 2
## USF1.1 4.523818e-07 0.7095087 0.045 0.114 1.655808e-02 2
## GTF2B 4.534235e-07 1.1257192 0.195 0.362 1.659621e-02 2
## ZNF738.1 4.570859e-07 0.3727003 0.020 0.073 1.673026e-02 2
## SPC24.2 4.681942e-07 -0.7862577 0.005 0.046 1.713684e-02 2
## DLGAP4 4.684923e-07 0.6037817 0.053 0.126 1.714775e-02 2
## PI4KB 4.687519e-07 1.0534228 0.063 0.143 1.715726e-02 2
## VTI1A.1 4.728905e-07 2.0023697 0.328 0.326 1.730874e-02 2
## NAT14 4.737181e-07 -1.5833314 0.003 0.043 1.733903e-02 2
## KLHL7 4.762862e-07 0.6582814 0.038 0.102 1.743303e-02 2
## TRBV7-9.2 4.776593e-07 -2.5580698 0.029 0.083 1.748329e-02 2
## FASTKD1 4.789989e-07 0.7698517 0.032 0.092 1.753232e-02 2
## UBA7 4.802231e-07 0.9263892 0.071 0.154 1.757713e-02 2
## CCND3.1 4.823494e-07 0.4318792 0.392 0.721 1.765495e-02 2
## DNAJC10 4.830059e-07 0.9988739 0.089 0.183 1.767898e-02 2
## AIG1.2 4.859699e-07 -0.8372144 0.005 0.046 1.778747e-02 2
## LINC02611 4.878294e-07 -0.1182323 0.009 0.054 1.785553e-02 2
## PDIA5.1 4.952046e-07 0.1859172 0.021 0.075 1.812548e-02 2
## FBXL14 4.956715e-07 0.4601492 0.032 0.092 1.814257e-02 2
## NAGA.2 4.975621e-07 -0.1120171 0.020 0.072 1.821177e-02 2
## NR4A2.2 5.010330e-07 0.7997314 0.146 0.270 1.833881e-02 2
## CANT1 5.022527e-07 0.8275650 0.054 0.128 1.838345e-02 2
## PPCDC.1 5.040412e-07 0.7143498 0.039 0.104 1.844892e-02 2
## RPP25.2 5.077874e-07 -0.8091760 0.008 0.051 1.858604e-02 2
## GADD45B.2 5.153005e-07 0.4767277 0.421 0.733 1.886103e-02 2
## C6orf62 5.178311e-07 0.8752181 0.072 0.156 1.895366e-02 2
## VCPKMT 5.227171e-07 0.6916649 0.036 0.099 1.913249e-02 2
## ACBD6 5.330154e-07 0.9417638 0.167 0.308 1.950943e-02 2
## AFG3L2.2 5.335310e-07 0.8503822 0.072 0.156 1.952830e-02 2
## FCGRT.1 5.340047e-07 -5.3706484 0.000 0.037 1.954564e-02 2
## LTBP4 5.392822e-07 0.8605626 0.041 0.107 1.973881e-02 2
## AGFG2 5.417692e-07 1.0089496 0.053 0.125 1.982984e-02 2
## STX7 5.429439e-07 0.6651895 0.050 0.120 1.987283e-02 2
## PAPOLG 5.477027e-07 0.5957928 0.044 0.111 2.004701e-02 2
## NDEL1 5.523136e-07 0.6740869 0.084 0.174 2.021578e-02 2
## HECTD3 5.571438e-07 0.3159062 0.027 0.085 2.039258e-02 2
## PKNOX1 5.584780e-07 0.5134438 0.035 0.097 2.044141e-02 2
## RNF13 5.684183e-07 0.8531918 0.120 0.232 2.080525e-02 2
## MTG2.1 5.714360e-07 0.8378352 0.036 0.099 2.091570e-02 2
## C11orf21.2 5.717262e-07 0.3388601 0.023 0.077 2.092632e-02 2
## CSPP1.2 5.736818e-07 0.9417969 0.057 0.132 2.099790e-02 2
## CLP1 5.780722e-07 -1.0433281 0.008 0.051 2.115860e-02 2
## GPBP1L1 5.793188e-07 0.8782104 0.120 0.232 2.120423e-02 2
## C21orf58.2 5.794868e-07 -0.2805843 0.015 0.064 2.121038e-02 2
## UBA6 5.800497e-07 0.9121627 0.126 0.241 2.123098e-02 2
## CDC25A.2 5.827815e-07 -1.0229132 0.003 0.043 2.133097e-02 2
## SUPT6H 5.842895e-07 0.9613173 0.131 0.249 2.138616e-02 2
## NBPF14.1 5.871711e-07 1.0544821 0.048 0.118 2.149164e-02 2
## GHDC.1 5.921719e-07 -0.4870522 0.011 0.056 2.167468e-02 2
## SEC14L1 5.925525e-07 0.9436215 0.093 0.188 2.168861e-02 2
## KLHL12 5.938864e-07 0.3352660 0.024 0.079 2.173743e-02 2
## DNAJB9 5.975629e-07 0.9240402 0.129 0.246 2.187200e-02 2
## CCP110.1 5.985675e-07 0.6859560 0.033 0.094 2.190877e-02 2
## EIF4E3.1 6.020031e-07 1.0177734 0.147 0.276 2.203452e-02 2
## MAP3K11 6.048362e-07 1.0710585 0.068 0.148 2.213821e-02 2
## ERO1A.1 6.079208e-07 0.9574572 0.205 0.375 2.225112e-02 2
## ODF3B.2 6.082268e-07 0.2033824 0.012 0.059 2.226232e-02 2
## FEM1B 6.125861e-07 0.5303442 0.039 0.103 2.242188e-02 2
## TRIP11 6.155626e-07 0.9850127 0.129 0.246 2.253082e-02 2
## ZWILCH.2 6.182253e-07 0.7327804 0.020 0.072 2.262828e-02 2
## CRAT 6.186749e-07 -0.9105610 0.008 0.051 2.264474e-02 2
## NEMF.1 6.216922e-07 0.9804381 0.188 0.345 2.275518e-02 2
## EFL1.1 6.229469e-07 0.8582805 0.060 0.137 2.280110e-02 2
## MCAT.1 6.230271e-07 -1.7597504 0.008 0.051 2.280404e-02 2
## TRBV28 6.249599e-07 -2.6135800 0.008 0.050 2.287478e-02 2
## SPINK2.2 6.297607e-07 -0.7372866 0.009 0.054 2.305050e-02 2
## NFYA 6.304281e-07 0.4801078 0.039 0.103 2.307493e-02 2
## DLEU1 6.320919e-07 1.2253647 0.068 0.148 2.313583e-02 2
## DNAJC11 6.511629e-07 0.9280520 0.048 0.118 2.383386e-02 2
## TAF11.1 6.531257e-07 0.9184462 0.060 0.136 2.390571e-02 2
## MCEE 6.579628e-07 0.6117523 0.038 0.101 2.408276e-02 2
## BBS7 6.705984e-07 0.4778218 0.033 0.094 2.454524e-02 2
## SHQ1 6.712255e-07 0.2956567 0.029 0.086 2.456819e-02 2
## ARHGAP5 6.715935e-07 1.1278350 0.075 0.160 2.458167e-02 2
## TOR1AIP2 6.748466e-07 0.6684789 0.066 0.145 2.470074e-02 2
## IER5L.2 6.763705e-07 0.9338399 0.077 0.162 2.475651e-02 2
## ITFG2 6.785963e-07 0.8925957 0.092 0.185 2.483798e-02 2
## AC111182.1.1 6.890013e-07 -0.8608835 0.011 0.056 2.521883e-02 2
## WDR83.1 6.946217e-07 0.6233825 0.032 0.091 2.542454e-02 2
## SPTY2D1 6.978404e-07 0.8049240 0.080 0.167 2.554235e-02 2
## RCBTB2.1 7.090946e-07 0.6677379 0.063 0.140 2.595428e-02 2
## SMAP1.1 7.134940e-07 0.8862622 0.233 0.424 2.611531e-02 2
## COQ10B 7.208207e-07 1.0354222 0.108 0.211 2.638348e-02 2
## BISPR.1 7.233385e-07 0.3526426 0.021 0.074 2.647564e-02 2
## SNRNP48 7.237848e-07 0.6478124 0.027 0.084 2.649197e-02 2
## MRPL1.1 7.266157e-07 0.8240849 0.066 0.145 2.659559e-02 2
## AL136454.1 7.281079e-07 -2.6358514 0.002 0.039 2.665021e-02 2
## CUL4A 7.324789e-07 1.0420384 0.150 0.280 2.681019e-02 2
## HIBADH.1 7.325479e-07 0.8431115 0.056 0.129 2.681272e-02 2
## DTD2 7.431171e-07 0.2817405 0.018 0.069 2.719957e-02 2
## GRPEL2 7.462225e-07 0.2222131 0.023 0.076 2.731324e-02 2
## GABPA 7.584924e-07 0.6384646 0.051 0.122 2.776234e-02 2
## ING2.1 7.707432e-07 0.1219572 0.017 0.066 2.821074e-02 2
## OGDH 7.710202e-07 0.9713239 0.144 0.270 2.822088e-02 2
## TROAP.2 7.749831e-07 -0.9959014 0.006 0.047 2.836593e-02 2
## MRI1.1 7.790117e-07 0.6903802 0.051 0.122 2.851339e-02 2
## AC139720.1.2 7.861682e-07 1.0107834 0.096 0.191 2.877533e-02 2
## MTFR2.2 7.903945e-07 0.7143739 0.009 0.053 2.893002e-02 2
## FLNB.1 7.925377e-07 0.7691333 0.041 0.105 2.900846e-02 2
## POLH.1 7.957563e-07 0.9747969 0.084 0.173 2.912627e-02 2
## VARS.1 8.000428e-07 0.9460190 0.050 0.119 2.928317e-02 2
## TUBGCP3.1 8.044287e-07 0.8778651 0.075 0.159 2.944370e-02 2
## CDPF1.1 8.047281e-07 0.3988309 0.018 0.069 2.945466e-02 2
## VSIG1.1 8.089854e-07 -0.9306878 0.008 0.050 2.961048e-02 2
## MTX2.1 8.314870e-07 0.8097459 0.065 0.143 3.043409e-02 2
## DPYD-AS1.2 8.332709e-07 0.5101812 0.042 0.107 3.049938e-02 2
## BCL2L1.2 8.349690e-07 1.1811900 0.129 0.246 3.056154e-02 2
## SCAF11 8.351842e-07 0.8392027 0.259 0.474 3.056941e-02 2
## POR 8.396712e-07 0.6499437 0.045 0.112 3.073364e-02 2
## DBN1.2 8.422987e-07 -1.1762095 0.008 0.050 3.082982e-02 2
## RNF125.1 8.434022e-07 0.8702854 0.238 0.431 3.087021e-02 2
## ZNF550 8.527265e-07 -0.3604306 0.012 0.058 3.121150e-02 2
## PCSK7.1 8.549054e-07 0.9577984 0.153 0.285 3.129125e-02 2
## NEMP2.1 8.554637e-07 0.3090098 0.020 0.071 3.131168e-02 2
## ATXN3 8.586716e-07 0.9537390 0.063 0.140 3.142910e-02 2
## LINC01184.2 8.594892e-07 1.1596284 0.077 0.161 3.145902e-02 2
## HERC1 8.596200e-07 1.8264853 0.370 0.396 3.146381e-02 2
## ENTPD6 8.613005e-07 0.6382412 0.024 0.078 3.152532e-02 2
## PIK3R2 8.616492e-07 0.4861266 0.020 0.071 3.153808e-02 2
## GZMH.2 8.705630e-07 -1.4215653 0.021 0.072 3.186435e-02 2
## AC119396.1.1 8.731812e-07 0.9272437 0.135 0.254 3.196018e-02 2
## RHOT1.1 8.761834e-07 1.0337560 0.057 0.130 3.207006e-02 2
## XRCC2.2 8.860031e-07 -0.4930846 0.003 0.041 3.242949e-02 2
## CCND2.1 8.862770e-07 0.6659828 0.298 0.542 3.243951e-02 2
## PACC1 8.875036e-07 0.6905591 0.041 0.105 3.248441e-02 2
## SLC25A51 8.897873e-07 0.3055495 0.039 0.102 3.256799e-02 2
## IRF2BP2 8.899706e-07 0.9957796 0.087 0.177 3.257471e-02 2
## C6orf120 8.965234e-07 0.6055157 0.029 0.086 3.281455e-02 2
## GGCX.1 8.980651e-07 -0.2249783 0.009 0.053 3.287098e-02 2
## MTRR 8.981414e-07 0.8213930 0.047 0.114 3.287377e-02 2
## BCL2L13 9.040269e-07 0.8253867 0.078 0.163 3.308919e-02 2
## ZNF160 9.056176e-07 1.0124975 0.075 0.159 3.314741e-02 2
## PSMA3-AS1 9.186119e-07 0.8943655 0.060 0.135 3.362303e-02 2
## STX6 9.229139e-07 0.6728038 0.059 0.133 3.378050e-02 2
## FAM118A.1 9.233014e-07 1.0115889 0.111 0.213 3.379468e-02 2
## UXS1.1 9.256197e-07 0.9629366 0.116 0.222 3.387953e-02 2
## PUS1.1 9.258413e-07 0.7541322 0.035 0.095 3.388764e-02 2
## HES6.2 9.354425e-07 -4.9722035 0.000 0.035 3.423907e-02 2
## VPS45 9.363283e-07 0.8058081 0.059 0.132 3.427149e-02 2
## CPSF6 9.437529e-07 0.8413200 0.167 0.307 3.454325e-02 2
## FIZ1.1 9.502030e-07 -1.1705132 0.003 0.041 3.477933e-02 2
## LINC00426 9.528139e-07 0.9537253 0.099 0.196 3.487489e-02 2
## EID3 9.562941e-07 -0.6361715 0.009 0.052 3.500228e-02 2
## EMILIN2.1 9.583288e-07 0.8269505 0.023 0.076 3.507675e-02 2
## CCNT1.1 9.617259e-07 1.0490282 0.087 0.177 3.520109e-02 2
## ZNF346 9.676696e-07 0.1996605 0.023 0.075 3.541864e-02 2
## EFCAB11.2 9.687764e-07 0.2310985 0.006 0.047 3.545915e-02 2
## ENOSF1.2 9.700351e-07 0.8585279 0.093 0.186 3.550523e-02 2
## RIOX1 9.754697e-07 -0.2803325 0.021 0.073 3.570414e-02 2
## CUL2 9.802505e-07 1.0462285 0.095 0.189 3.587913e-02 2
## PSD4.2 9.830834e-07 0.9701573 0.131 0.247 3.598282e-02 2
## VILL 9.873723e-07 0.5984687 0.021 0.073 3.613980e-02 2
## MED21.1 9.886861e-07 0.7934242 0.042 0.107 3.618789e-02 2
## DIS3 9.908925e-07 0.7868259 0.071 0.151 3.626865e-02 2
## KRBOX4 9.915802e-07 0.3993837 0.030 0.087 3.629382e-02 2
## TGOLN2.1 9.944580e-07 0.9746720 0.188 0.343 3.639915e-02 2
## HGS 9.992909e-07 1.0428601 0.084 0.172 3.657605e-02 2
## LRRC40.2 1.003604e-06 0.6447202 0.044 0.109 3.673392e-02 2
## PYGO2 1.003942e-06 0.7172651 0.036 0.097 3.674629e-02 2
## CABIN1 1.006954e-06 1.0722546 0.162 0.300 3.685653e-02 2
## RNF44 1.016081e-06 0.8347938 0.086 0.175 3.719059e-02 2
## SH3BP5L 1.036404e-06 -0.5002840 0.014 0.060 3.793445e-02 2
## BTN2A1 1.037186e-06 0.8376982 0.062 0.137 3.796309e-02 2
## FAM189B 1.037196e-06 -0.5580897 0.006 0.047 3.796346e-02 2
## C4orf46.2 1.038515e-06 -0.7830617 0.005 0.044 3.801173e-02 2
## ZNF302.1 1.055076e-06 0.7630432 0.027 0.083 3.861788e-02 2
## TOMM34.1 1.060615e-06 0.4050534 0.029 0.085 3.882064e-02 2
## RNFT1 1.063590e-06 0.4034993 0.032 0.090 3.892952e-02 2
## SNHG12 1.066016e-06 1.0699689 0.132 0.248 3.901831e-02 2
## TBK1.1 1.068212e-06 0.6847683 0.093 0.186 3.909869e-02 2
## EEPD1 1.085928e-06 -0.2535719 0.009 0.052 3.974714e-02 2
## WDR86-AS1.1 1.098344e-06 -1.6274912 0.008 0.049 4.020160e-02 2
## LRRCC1.2 1.101097e-06 -0.4242098 0.006 0.047 4.030235e-02 2
## MCCC1 1.106638e-06 0.7371562 0.054 0.125 4.050518e-02 2
## HYI.2 1.106833e-06 0.2491593 0.009 0.052 4.051231e-02 2
## C7orf25 1.108086e-06 -0.6823840 0.006 0.047 4.055817e-02 2
## TPM4.2 1.110441e-06 0.6079467 0.320 0.574 4.064437e-02 2
## ZNF692 1.112019e-06 0.8578663 0.027 0.083 4.070210e-02 2
## UBE2K 1.112846e-06 0.9654996 0.182 0.332 4.073239e-02 2
## BAZ2A 1.127274e-06 1.0504160 0.146 0.270 4.126049e-02 2
## C3orf38 1.139801e-06 0.5327621 0.048 0.115 4.171898e-02 2
## MTA1.1 1.142127e-06 0.9436657 0.051 0.120 4.180413e-02 2
## ZNF683.2 1.142333e-06 -0.3598635 0.051 0.117 4.181166e-02 2
## UPRT 1.146382e-06 -0.2172312 0.021 0.072 4.195989e-02 2
## ODC1 1.151468e-06 1.1507098 0.226 0.406 4.214604e-02 2
## USP21 1.157027e-06 0.5710426 0.035 0.094 4.234951e-02 2
## PBK.2 1.166714e-06 -0.8720386 0.003 0.041 4.270405e-02 2
## ZFP90 1.167821e-06 0.7553212 0.042 0.106 4.274458e-02 2
## RILP.1 1.169786e-06 -1.2704457 0.003 0.041 4.281650e-02 2
## STRADA 1.183740e-06 0.9016807 0.065 0.142 4.332725e-02 2
## MSANTD3 1.184399e-06 -0.5712903 0.011 0.055 4.335137e-02 2
## PAX8-AS1 1.186690e-06 -0.3367343 0.015 0.062 4.343522e-02 2
## PCF11 1.191981e-06 0.9535399 0.108 0.209 4.362887e-02 2
## LONP1.2 1.197650e-06 1.1466830 0.045 0.111 4.383638e-02 2
## SGSH.2 1.218581e-06 0.2024405 0.026 0.080 4.460250e-02 2
## LINS1.1 1.234717e-06 1.0331734 0.104 0.202 4.519310e-02 2
## OSBPL3.1 1.237307e-06 2.2089780 0.290 0.271 4.528789e-02 2
## SLC30A9 1.237917e-06 0.8771259 0.104 0.201 4.531022e-02 2
## RAD54B.2 1.260231e-06 -0.9553039 0.006 0.046 4.612697e-02 2
## FAM24B 1.261354e-06 -0.6040150 0.015 0.062 4.616808e-02 2
## ADAT3.1 1.265996e-06 -0.8355980 0.006 0.046 4.633800e-02 2
## TTI1.1 1.267866e-06 0.6827321 0.044 0.108 4.640642e-02 2
## APTR 1.268887e-06 -1.4846272 0.005 0.043 4.644379e-02 2
## TMEM185A 1.287300e-06 0.3624770 0.026 0.079 4.711777e-02 2
## NFATC2IP.1 1.298023e-06 1.1492609 0.168 0.306 4.751023e-02 2
## THAP3 1.302698e-06 0.6657648 0.023 0.075 4.768134e-02 2
## RNMT 1.304021e-06 1.0817782 0.116 0.221 4.772978e-02 2
## VAPB.1 1.326473e-06 0.8939106 0.057 0.129 4.855158e-02 2
## C5orf56 1.329349e-06 0.9609530 0.189 0.342 4.865684e-02 2
## ZNF264 1.337607e-06 0.5079202 0.035 0.094 4.895909e-02 2
## MXRA7.1 1.347030e-06 0.6771467 0.020 0.070 4.930398e-02 2
## TYK2 1.357496e-06 1.1946914 0.062 0.136 4.968705e-02 2
## YAE1 1.360063e-06 -0.8533892 0.006 0.046 4.978102e-02 2
## CEP97.1 1.365190e-06 0.8602832 0.038 0.099 4.996868e-02 2
## NSDHL.1 1.370405e-06 0.2837033 0.014 0.059 5.015957e-02 2
## NHEJ1 1.386593e-06 0.9640584 0.036 0.096 5.075209e-02 2
## SNRNP200 1.391149e-06 1.1041484 0.170 0.310 5.091884e-02 2
## UGDH 1.401019e-06 -0.5524898 0.015 0.062 5.128008e-02 2
## ZMYM6 1.405900e-06 0.9899010 0.060 0.134 5.145874e-02 2
## SLC38A2 1.417400e-06 2.0374516 0.415 0.484 5.187966e-02 2
## UBQLN1 1.422687e-06 0.9042533 0.149 0.274 5.207319e-02 2
## BIK 1.423761e-06 -5.1430241 0.000 0.034 5.211250e-02 2
## FOSB 1.445194e-06 1.4594224 0.433 0.501 5.289698e-02 2
## PATZ1 1.449956e-06 0.2792417 0.029 0.084 5.307130e-02 2
## PRR34-AS1 1.464005e-06 -2.5841474 0.002 0.037 5.358550e-02 2
## MED7 1.473745e-06 1.1754723 0.030 0.087 5.394202e-02 2
## TCF3.1 1.479365e-06 1.2422004 0.044 0.108 5.414773e-02 2
## SMARCA4.1 1.484866e-06 1.1456283 0.135 0.253 5.434907e-02 2
## EAF1.2 1.490763e-06 0.5329124 0.041 0.103 5.456491e-02 2
## NNT-AS1 1.493935e-06 0.1072087 0.018 0.067 5.468101e-02 2
## APOOL 1.495622e-06 0.2027772 0.021 0.072 5.474274e-02 2
## ZNF611 1.505927e-06 0.7183206 0.054 0.124 5.511993e-02 2
## GRK2.1 1.522861e-06 0.9569560 0.230 0.415 5.573976e-02 2
## MAK16 1.525938e-06 0.4112213 0.018 0.067 5.585238e-02 2
## PPP1R21 1.533615e-06 1.0039625 0.101 0.197 5.613339e-02 2
## ARAP2.2 1.552082e-06 1.9164202 0.356 0.377 5.680931e-02 2
## RTL6 1.572188e-06 -1.3394854 0.005 0.043 5.754521e-02 2
## FKBP2 1.572757e-06 -1.3066663 0.005 0.043 5.756604e-02 2
## PHIP.1 1.613931e-06 0.9377187 0.220 0.394 5.907312e-02 2
## IL3RA 1.614978e-06 -0.4997027 0.009 0.051 5.911142e-02 2
## AGAP2 1.623718e-06 0.9280339 0.078 0.161 5.943131e-02 2
## INTS9 1.660355e-06 0.8071805 0.050 0.117 6.077231e-02 2
## ADAM8.2 1.675218e-06 0.9700051 0.167 0.303 6.131634e-02 2
## ADAR 1.687637e-06 0.9395099 0.233 0.420 6.177090e-02 2
## WDR89 1.697824e-06 0.4349664 0.027 0.081 6.214375e-02 2
## C9orf85 1.711065e-06 0.7934494 0.054 0.123 6.262841e-02 2
## DEPDC1B.2 1.711114e-06 -0.1760195 0.009 0.051 6.263019e-02 2
## ZNF426 1.714684e-06 0.3988619 0.021 0.071 6.276086e-02 2
## PCED1B.2 1.716467e-06 0.9142480 0.211 0.378 6.282612e-02 2
## TXNRD2.1 1.722176e-06 0.1985129 0.023 0.074 6.303508e-02 2
## TMCO6 1.724856e-06 0.5284323 0.018 0.066 6.313318e-02 2
## DIABLO 1.752794e-06 0.1957240 0.015 0.061 6.415578e-02 2
## FHL3 1.755723e-06 -2.5502374 0.003 0.039 6.426296e-02 2
## RNF149.1 1.764932e-06 0.9493385 0.253 0.457 6.460004e-02 2
## NFATC1.1 1.766923e-06 1.0241001 0.063 0.137 6.467293e-02 2
## TTC5 1.780268e-06 0.7015417 0.035 0.093 6.516136e-02 2
## MRE11.1 1.782506e-06 0.8373081 0.075 0.156 6.524329e-02 2
## LARP1.2 1.786574e-06 1.0089314 0.120 0.227 6.539217e-02 2
## SMAD4 1.786626e-06 0.8677990 0.116 0.220 6.539410e-02 2
## NSUN2 1.811408e-06 0.8791626 0.110 0.210 6.630114e-02 2
## ZUP1 1.820137e-06 0.4605957 0.035 0.093 6.662066e-02 2
## LUC7L2.1 1.833774e-06 0.8819976 0.253 0.459 6.711981e-02 2
## SNIP1.1 1.855741e-06 1.0124821 0.057 0.128 6.792382e-02 2
## TEDC2.2 1.899695e-06 -0.8009717 0.003 0.039 6.953265e-02 2
## B4GAT1 1.902544e-06 -1.3240310 0.003 0.039 6.963692e-02 2
## CTC1 1.924177e-06 0.8888778 0.066 0.141 7.042874e-02 2
## NUFIP1 1.925659e-06 0.4879744 0.030 0.085 7.048299e-02 2
## SPATS2.1 1.933413e-06 0.8017011 0.038 0.097 7.076677e-02 2
## PPM1A 1.937676e-06 0.7092710 0.060 0.132 7.092282e-02 2
## SAMD3.2 1.938110e-06 0.9369780 0.217 0.388 7.093871e-02 2
## ZNF226 1.947879e-06 0.1074495 0.021 0.071 7.129627e-02 2
## CCDC9 1.950983e-06 0.6032958 0.026 0.078 7.140986e-02 2
## SLC25A45 1.959676e-06 0.6841403 0.042 0.104 7.172805e-02 2
## LTB4R2.1 1.965569e-06 -0.5226526 0.011 0.053 7.194376e-02 2
## XPO5 1.968791e-06 0.5408947 0.045 0.109 7.206169e-02 2
## CSF1.2 1.989905e-06 0.3157306 0.048 0.113 7.283452e-02 2
## RAB34 2.015128e-06 -1.3390828 0.006 0.045 7.375771e-02 2
## RCAN1 2.020686e-06 -0.1795412 0.011 0.053 7.396113e-02 2
## SLC18B1 2.039326e-06 -1.4669353 0.003 0.039 7.464342e-02 2
## IZUMO4 2.057109e-06 0.1902312 0.006 0.045 7.529432e-02 2
## MOSPD2 2.061011e-06 0.7847573 0.036 0.095 7.543711e-02 2
## RIBC2.2 2.076323e-06 -1.4633596 0.002 0.036 7.599756e-02 2
## SMC5.1 2.079149e-06 1.0705279 0.183 0.331 7.610100e-02 2
## ZNF7.1 2.079314e-06 0.3148021 0.024 0.076 7.610705e-02 2
## RPE 2.083966e-06 0.6300199 0.033 0.090 7.627733e-02 2
## SCPEP1.1 2.097213e-06 -0.1490813 0.012 0.055 7.676218e-02 2
## SLC27A3.1 2.101702e-06 0.6305695 0.024 0.075 7.692650e-02 2
## HEATR6 2.109702e-06 0.9194577 0.039 0.099 7.721930e-02 2
## RNF135 2.131976e-06 -0.1746738 0.015 0.061 7.803458e-02 2
## PRIMPOL.1 2.132582e-06 0.4887113 0.030 0.085 7.805675e-02 2
## NAT9 2.146081e-06 0.4262731 0.027 0.080 7.855085e-02 2
## TRAV17.1 2.152166e-06 -1.4565649 0.017 0.062 7.877358e-02 2
## AP000787.1.1 2.157672e-06 0.3467219 0.026 0.078 7.897512e-02 2
## RYK 2.186031e-06 0.5095155 0.036 0.095 8.001310e-02 2
## ARHGEF1 2.189300e-06 1.3469198 0.484 0.645 8.013274e-02 2
## PLK1.2 2.192361e-06 -1.4917666 0.005 0.042 8.024481e-02 2
## ERGIC1.1 2.215988e-06 0.7833439 0.048 0.113 8.110959e-02 2
## PPM1D 2.220454e-06 0.9858116 0.054 0.122 8.127306e-02 2
## AMPD2 2.222838e-06 0.7807273 0.036 0.095 8.136031e-02 2
## NAPA-AS1 2.223439e-06 -0.9999126 0.005 0.042 8.138233e-02 2
## ZNF397 2.230193e-06 -0.3967405 0.012 0.055 8.162952e-02 2
## PIK3R1.1 2.248007e-06 0.9051114 0.254 0.460 8.228155e-02 2
## FKBP15.1 2.261543e-06 1.1247613 0.062 0.134 8.277701e-02 2
## ZEB1.1 2.263343e-06 1.7475832 0.385 0.434 8.284289e-02 2
## TM9SF4 2.268775e-06 0.9523536 0.069 0.145 8.304169e-02 2
## TNFRSF18.1 2.270889e-06 -0.5508475 0.018 0.065 8.311908e-02 2
## NFKBIA.1 2.302835e-06 0.6946706 0.337 0.592 8.428837e-02 2
## SHOC2 2.305272e-06 0.8301730 0.099 0.192 8.437758e-02 2
## IPO4.1 2.317847e-06 0.3747048 0.024 0.075 8.483785e-02 2
## VSIG8 2.318031e-06 4.4196225 0.017 0.003 8.484457e-02 2
## ARHGEF2 2.324335e-06 0.9171034 0.090 0.178 8.507529e-02 2
## TNFAIP3 2.349868e-06 1.4660348 0.483 0.593 8.600988e-02 2
## VEZT 2.353343e-06 0.7940098 0.083 0.166 8.613707e-02 2
## AASDH.1 2.374665e-06 0.7051623 0.021 0.070 8.691750e-02 2
## WDR6 2.374772e-06 1.0898277 0.074 0.153 8.692140e-02 2
## B4GALT7 2.377556e-06 0.1874937 0.026 0.078 8.702331e-02 2
## PARP11.2 2.387048e-06 0.7510253 0.059 0.129 8.737074e-02 2
## GTF3C3 2.420496e-06 0.7912663 0.048 0.113 8.859498e-02 2
## COQ10A 2.421259e-06 0.3689211 0.030 0.085 8.862291e-02 2
## CTPS1.2 2.423132e-06 1.0980549 0.068 0.143 8.869149e-02 2
## CA5B 2.424095e-06 0.8514523 0.054 0.122 8.872672e-02 2
## SAP130.1 2.427825e-06 1.0963824 0.059 0.129 8.886324e-02 2
## FAM120A 2.430961e-06 0.9634123 0.171 0.311 8.897802e-02 2
## RNF2 2.433875e-06 0.7002550 0.042 0.103 8.908469e-02 2
## UBAP2.1 2.446968e-06 1.0493408 0.089 0.175 8.956392e-02 2
## PCBP4.1 2.455228e-06 0.3100973 0.017 0.063 8.986626e-02 2
## UFSP2 2.456207e-06 0.8154868 0.036 0.094 8.990208e-02 2
## TC2N.2 2.475761e-06 1.2972646 0.490 0.641 9.061779e-02 2
## CCDC14.1 2.478662e-06 0.9479818 0.081 0.164 9.072400e-02 2
## UBE4A 2.487107e-06 0.7698526 0.110 0.208 9.103309e-02 2
## ZBTB4 2.487844e-06 0.4942114 0.044 0.106 9.106007e-02 2
## ZNF33B.1 2.488765e-06 0.6778082 0.053 0.120 9.109379e-02 2
## HELZ 2.493732e-06 1.9461899 0.326 0.332 9.127558e-02 2
## WDR4.2 2.496825e-06 0.8204520 0.027 0.080 9.138878e-02 2
## SEC23A.1 2.502286e-06 0.9997256 0.057 0.127 9.158867e-02 2
## APTX 2.519358e-06 0.7606788 0.080 0.160 9.221355e-02 2
## MALT1 2.535988e-06 1.2421887 0.147 0.270 9.282223e-02 2
## FECH.1 2.541521e-06 0.2254856 0.014 0.058 9.302473e-02 2
## GABARAPL1.2 2.545420e-06 0.8815762 0.071 0.148 9.316748e-02 2
## INTS3 2.548431e-06 1.0800028 0.045 0.108 9.327769e-02 2
## LY9 2.557043e-06 0.2274620 0.033 0.089 9.359289e-02 2
## TRAV8-4 2.565444e-06 -1.7385938 0.008 0.047 9.390039e-02 2
## TADA2A.2 2.603099e-06 0.7291680 0.036 0.094 9.527865e-02 2
## ELAC1 2.606684e-06 -0.2610152 0.017 0.063 9.540983e-02 2
## ZNF322 2.612723e-06 0.4387757 0.020 0.067 9.563089e-02 2
## EEF1AKMT4 2.635805e-06 -0.4991039 0.009 0.050 9.647573e-02 2
## MARK2 2.639713e-06 0.9596172 0.102 0.196 9.661878e-02 2
## ZDHHC7 2.654612e-06 0.7560768 0.038 0.096 9.716410e-02 2
## ATP2A1-AS1.2 2.671801e-06 -4.7815788 0.000 0.032 9.779325e-02 2
## PDCD11 2.687953e-06 0.8864892 0.045 0.108 9.838444e-02 2
## IDE 2.700944e-06 0.7344148 0.059 0.129 9.885996e-02 2
## TADA2B 2.709801e-06 0.3392960 0.029 0.082 9.918414e-02 2
## KIF14.2 2.721081e-06 -0.6832833 0.008 0.047 9.959701e-02 2
## ACYP2 2.728578e-06 1.3733213 0.158 0.288 9.987141e-02 2
## IPO8 2.730718e-06 0.6728038 0.057 0.126 9.994975e-02 2
## RBM10 2.756845e-06 0.9997018 0.078 0.158 1.009061e-01 2
## CDC16 2.783018e-06 0.8856188 0.102 0.196 1.018640e-01 2
## FANCM.2 2.795328e-06 0.5780300 0.023 0.072 1.023146e-01 2
## XPO7.1 2.807654e-06 1.0073124 0.069 0.145 1.027657e-01 2
## HCG18 2.828190e-06 1.0110236 0.089 0.174 1.035174e-01 2
## UEVLD.1 2.833729e-06 0.3316579 0.030 0.084 1.037201e-01 2
## FAM126A 2.852628e-06 0.9770263 0.116 0.218 1.044119e-01 2
## AC125494.1 2.860404e-06 -2.4541482 0.003 0.038 1.046965e-01 2
## POLR3H.2 2.877344e-06 0.9392594 0.029 0.082 1.053165e-01 2
## ZBTB14 2.888665e-06 0.6549008 0.021 0.070 1.057309e-01 2
## ZBED5-AS1 2.889708e-06 -0.6955325 0.006 0.044 1.057691e-01 2
## UBASH3A.1 2.894520e-06 1.2099599 0.183 0.331 1.059452e-01 2
## SPPL2A.1 2.912456e-06 1.0526576 0.143 0.262 1.066017e-01 2
## EIPR1.1 2.939564e-06 1.1419819 0.099 0.191 1.075939e-01 2
## TIMM44.1 2.963596e-06 0.5630897 0.026 0.077 1.084735e-01 2
## SLC35A1 2.970605e-06 0.8512309 0.050 0.115 1.087301e-01 2
## TRMO 2.972735e-06 1.1173325 0.080 0.161 1.088081e-01 2
## TXNL4B 2.978961e-06 0.4920421 0.020 0.067 1.090359e-01 2
## LHFPL5 3.008918e-06 -0.2396479 0.009 0.049 1.101324e-01 2
## SNX12 3.042739e-06 -0.1498420 0.014 0.057 1.113703e-01 2
## GMFB 3.065343e-06 0.5992476 0.042 0.103 1.121977e-01 2
## PCYOX1 3.071558e-06 0.5949313 0.024 0.074 1.124252e-01 2
## ZC3HC1.1 3.077910e-06 0.7092218 0.027 0.079 1.126577e-01 2
## PPOX 3.087300e-06 0.8909591 0.041 0.101 1.130013e-01 2
## SNTA1.1 3.094518e-06 -0.4254787 0.014 0.057 1.132655e-01 2
## AC017083.2 3.095746e-06 -0.5060349 0.003 0.038 1.133105e-01 2
## MYPOP 3.129562e-06 -0.7219375 0.012 0.054 1.145482e-01 2
## CCDC130 3.137339e-06 1.1679548 0.086 0.170 1.148329e-01 2
## COQ8B 3.150657e-06 0.9129306 0.041 0.100 1.153204e-01 2
## MEPCE 3.167862e-06 0.8132591 0.045 0.107 1.159501e-01 2
## DPP9 3.226442e-06 1.2712273 0.089 0.174 1.180942e-01 2
## SCFD1 3.232143e-06 0.8831729 0.164 0.294 1.183029e-01 2
## SERPINB9.1 3.252629e-06 1.4553095 0.108 0.204 1.190527e-01 2
## DCTN4 3.255820e-06 0.9951805 0.105 0.200 1.191695e-01 2
## CEP295.2 3.269113e-06 0.9252516 0.057 0.125 1.196561e-01 2
## C11orf71 3.294128e-06 0.6296291 0.018 0.064 1.205717e-01 2
## HMMR.2 3.321124e-06 0.4407338 0.026 0.076 1.215598e-01 2
## INTS7.1 3.344668e-06 0.7236381 0.062 0.133 1.224215e-01 2
## FRMD8 3.357150e-06 0.8589516 0.045 0.107 1.228784e-01 2
## MOB3C 3.361299e-06 -0.2703342 0.014 0.057 1.230303e-01 2
## BCDIN3D 3.365055e-06 0.6733058 0.024 0.074 1.231677e-01 2
## GPAT4 3.379550e-06 0.5129822 0.036 0.093 1.236983e-01 2
## STRADB 3.379854e-06 0.4687198 0.026 0.076 1.237094e-01 2
## DDX55.2 3.383689e-06 1.0775433 0.054 0.121 1.238498e-01 2
## KIF9.1 3.384209e-06 0.5831059 0.035 0.091 1.238688e-01 2
## MSRA.1 3.387320e-06 0.9231375 0.056 0.123 1.239827e-01 2
## ZNF85.1 3.399973e-06 0.3460576 0.018 0.064 1.244458e-01 2
## PHYH 3.434594e-06 1.1209683 0.035 0.091 1.257130e-01 2
## NARF 3.505037e-06 1.4053464 0.137 0.251 1.282914e-01 2
## TP53BP1 3.520086e-06 1.0969572 0.089 0.174 1.288422e-01 2
## SCRN3 3.556714e-06 0.5293859 0.024 0.074 1.301828e-01 2
## TCAIM 3.572287e-06 0.5997611 0.051 0.116 1.307529e-01 2
## TCEAL4.1 3.599356e-06 -3.3571104 0.002 0.034 1.317436e-01 2
## TCOF1.1 3.619466e-06 0.9368329 0.071 0.146 1.324797e-01 2
## ZSCAN32 3.623761e-06 0.6487002 0.023 0.071 1.326369e-01 2
## NMB.1 3.624675e-06 -2.4357570 0.002 0.034 1.326704e-01 2
## SLC25A19.2 3.672151e-06 1.1139273 0.023 0.071 1.344081e-01 2
## TICAM1 3.678868e-06 0.9099661 0.042 0.102 1.346539e-01 2
## CEP290 3.686799e-06 0.9637950 0.066 0.139 1.349442e-01 2
## IQCB1.1 3.707820e-06 0.9313409 0.107 0.201 1.357136e-01 2
## PRKDC.2 3.713146e-06 0.7007790 0.244 0.428 1.359086e-01 2
## CAMK2N1.1 3.715531e-06 -1.5541769 0.006 0.043 1.359959e-01 2
## APOBEC3F 3.722436e-06 0.4599057 0.017 0.061 1.362486e-01 2
## KATNAL1 3.724055e-06 0.8217586 0.065 0.137 1.363079e-01 2
## ITPR2 3.770953e-06 1.9604075 0.344 0.364 1.380244e-01 2
## WDR74 3.775918e-06 1.2418412 0.173 0.310 1.382062e-01 2
## ANKRD9.2 3.789262e-06 -4.7341423 0.000 0.031 1.386946e-01 2
## MAN2B2 3.819794e-06 0.7655511 0.054 0.120 1.398121e-01 2
## IRF2BP1 3.841323e-06 -0.2166675 0.017 0.061 1.406001e-01 2
## FASTKD3 3.856054e-06 0.2654271 0.020 0.066 1.411393e-01 2
## C2CD2L.1 3.858965e-06 0.7074493 0.038 0.095 1.412459e-01 2
## HOMEZ 3.865983e-06 0.4487262 0.017 0.061 1.415027e-01 2
## TBC1D15 3.869694e-06 0.9446380 0.089 0.174 1.416386e-01 2
## PRKCD 3.874491e-06 0.4180116 0.026 0.076 1.418141e-01 2
## METTL6 3.875051e-06 0.5930991 0.039 0.097 1.418346e-01 2
## FAAP24.2 3.887851e-06 -2.1886216 0.002 0.034 1.423031e-01 2
## HRH2.2 3.887851e-06 -1.3134396 0.002 0.034 1.423031e-01 2
## C1GALT1.1 3.895911e-06 1.0214080 0.150 0.273 1.425981e-01 2
## R3HDM1.1 3.903187e-06 1.0412078 0.168 0.302 1.428644e-01 2
## ZNF280D 3.924746e-06 1.0022760 0.072 0.148 1.436535e-01 2
## AKAP17A 3.960973e-06 1.1415593 0.165 0.298 1.449795e-01 2
## RANBP6 3.985144e-06 0.7925741 0.047 0.109 1.458642e-01 2
## ODR4.1 3.993106e-06 0.7284160 0.053 0.118 1.461557e-01 2
## AC004797.1 4.000996e-06 0.2810902 0.014 0.056 1.464444e-01 2
## RNF139 4.018390e-06 0.8534268 0.086 0.169 1.470811e-01 2
## DDX11.2 4.064149e-06 1.0233131 0.051 0.115 1.487560e-01 2
## POT1 4.067090e-06 0.4339539 0.032 0.085 1.488636e-01 2
## RAD51D.1 4.075491e-06 0.5179185 0.030 0.083 1.491711e-01 2
## NUP43.1 4.092964e-06 0.7152115 0.024 0.073 1.498107e-01 2
## CRTC3.1 4.100830e-06 1.0952493 0.140 0.254 1.500986e-01 2
## POM121C 4.125137e-06 0.8202841 0.069 0.144 1.509883e-01 2
## LINC00476 4.149790e-06 0.9700115 0.069 0.143 1.518906e-01 2
## NFE2L3.2 4.157127e-06 0.9009391 0.078 0.157 1.521592e-01 2
## SP3.1 4.166791e-06 1.0497250 0.149 0.268 1.525129e-01 2
## SLC25A28 4.206068e-06 0.9431647 0.045 0.107 1.539505e-01 2
## ARHGEF6.1 4.210110e-06 0.9415439 0.223 0.398 1.540985e-01 2
## C16orf74.2 4.274598e-06 -1.4841401 0.006 0.043 1.564588e-01 2
## CHD7.1 4.276508e-06 1.0308974 0.068 0.141 1.565288e-01 2
## IL6ST.2 4.285284e-06 1.0289478 0.125 0.229 1.568500e-01 2
## PRKCI 4.305041e-06 0.7909789 0.039 0.097 1.575731e-01 2
## GOLPH3L 4.333899e-06 0.7353322 0.029 0.080 1.586294e-01 2
## PPP3R1.1 4.363218e-06 0.9625076 0.170 0.303 1.597025e-01 2
## NUP210.2 4.410363e-06 0.8950294 0.182 0.321 1.614281e-01 2
## GEMIN2.1 4.421523e-06 0.9393937 0.021 0.068 1.618366e-01 2
## C17orf58.1 4.422081e-06 -0.8406715 0.005 0.040 1.618570e-01 2
## GRIPAP1.1 4.449101e-06 1.4193430 0.108 0.204 1.628460e-01 2
## LINC01572.2 4.464214e-06 0.3158061 0.027 0.078 1.633992e-01 2
## ZNF71 4.468305e-06 -0.1763240 0.008 0.045 1.635489e-01 2
## ATG16L2 4.471277e-06 1.0827049 0.104 0.196 1.636577e-01 2
## TGFBRAP1 4.528352e-06 0.9319329 0.041 0.099 1.657468e-01 2
## DHX16.1 4.547338e-06 1.0664191 0.072 0.148 1.664417e-01 2
## HIGD1A 4.556703e-06 -0.5611765 0.009 0.048 1.667844e-01 2
## GUF1 4.558686e-06 0.7669684 0.051 0.115 1.668570e-01 2
## PIGU.2 4.573101e-06 0.5095800 0.032 0.085 1.673846e-01 2
## NUP205.2 4.592013e-06 1.0522396 0.065 0.136 1.680768e-01 2
## PCYT1A.1 4.592747e-06 1.1031955 0.098 0.187 1.681037e-01 2
## CNOT2 4.595274e-06 0.9245054 0.198 0.353 1.681962e-01 2
## FBXL17.1 4.599037e-06 2.1849331 0.298 0.290 1.683339e-01 2
## NEK9 4.603281e-06 0.6679586 0.068 0.140 1.684893e-01 2
## PLAGL2.1 4.603796e-06 0.3871183 0.015 0.058 1.685081e-01 2
## TMEM63B 4.622248e-06 -0.8461742 0.006 0.043 1.691835e-01 2
## MLLT11.1 4.652696e-06 0.4985088 0.021 0.068 1.702980e-01 2
## NUDT15.1 4.653334e-06 0.6343145 0.021 0.068 1.703213e-01 2
## ACADSB 4.657257e-06 0.5850243 0.033 0.087 1.704649e-01 2
## AP002360.1 4.670937e-06 -0.7632844 0.005 0.040 1.709656e-01 2
## LPCAT3 4.714496e-06 0.8778788 0.063 0.134 1.725600e-01 2
## SNRPD3.1 4.743518e-06 -0.8599034 0.005 0.040 1.736222e-01 2
## NDUFAF6.1 4.751919e-06 1.2931423 0.078 0.157 1.739297e-01 2
## ATRIP.1 4.758345e-06 0.6677813 0.021 0.068 1.741649e-01 2
## GFOD2 4.758479e-06 0.6009249 0.017 0.061 1.741698e-01 2
## CSNK1D 4.774836e-06 1.0717521 0.126 0.232 1.747685e-01 2
## RANBP3.1 4.830295e-06 0.9694180 0.063 0.134 1.767985e-01 2
## CCDC138.1 4.833705e-06 0.3875603 0.035 0.089 1.769233e-01 2
## TRAV29DV5.1 4.837467e-06 -1.1508570 0.027 0.076 1.770610e-01 2
## ALG6 4.904127e-06 0.8067712 0.041 0.099 1.795008e-01 2
## PUM3.1 4.921296e-06 1.1537317 0.105 0.198 1.801293e-01 2
## NACC1.1 4.947303e-06 1.1103312 0.030 0.083 1.810812e-01 2
## ALKBH4 4.971005e-06 1.0530292 0.033 0.087 1.819487e-01 2
## MICALL1 4.999172e-06 0.3229621 0.029 0.080 1.829797e-01 2
## PBX2 5.012345e-06 0.5521415 0.029 0.080 1.834619e-01 2
## HYAL3 5.028484e-06 -1.4055900 0.003 0.037 1.840526e-01 2
## NT5C2 5.050461e-06 0.9996841 0.122 0.223 1.848570e-01 2
## AL391056.1.1 5.064582e-06 -1.2965318 0.005 0.039 1.853738e-01 2
## ACIN1 5.083142e-06 1.0303495 0.220 0.389 1.860532e-01 2
## BEST1 5.094065e-06 0.7993337 0.026 0.075 1.864530e-01 2
## TARS2.1 5.113021e-06 0.7771239 0.041 0.099 1.871468e-01 2
## RNF26 5.141558e-06 0.1674151 0.011 0.050 1.881913e-01 2
## SIN3B 5.148427e-06 0.9516934 0.038 0.094 1.884427e-01 2
## POLR2J3 5.150277e-06 1.1246833 0.129 0.236 1.885104e-01 2
## LMBR1L 5.154759e-06 0.7620652 0.036 0.092 1.886745e-01 2
## AC005332.4 5.188192e-06 0.5716529 0.032 0.085 1.898982e-01 2
## HIST1H3G.2 5.209851e-06 -0.3922953 0.012 0.053 1.906910e-01 2
## EIF4G3.1 5.222119e-06 1.8853991 0.328 0.336 1.911400e-01 2
## FBXO8 5.269951e-06 0.3464120 0.021 0.068 1.928907e-01 2
## TUBGCP4 5.286117e-06 0.9187983 0.027 0.078 1.934825e-01 2
## SENP3 5.286328e-06 0.6697291 0.023 0.070 1.934902e-01 2
## ZC3H11A 5.320494e-06 -0.2936754 0.012 0.053 1.947407e-01 2
## C9orf135.2 5.363692e-06 -0.5078942 0.011 0.050 1.963219e-01 2
## REG4.1 5.373186e-06 -5.6264929 0.000 0.030 1.966693e-01 2
## GABPB1-AS1 5.397696e-06 1.0235877 0.117 0.216 1.975665e-01 2
## TONSL.2 5.401500e-06 -0.5580281 0.005 0.039 1.977057e-01 2
## TRMT13 5.424764e-06 0.9971798 0.036 0.092 1.985572e-01 2
## CENPQ.2 5.448391e-06 -0.3484697 0.008 0.045 1.994220e-01 2
## ABITRAM 5.481251e-06 -0.1808872 0.014 0.055 2.006248e-01 2
## MASTL.2 5.496407e-06 0.7839652 0.032 0.084 2.011795e-01 2
## SULT1A1.1 5.513978e-06 -0.3503599 0.015 0.058 2.018226e-01 2
## AC016747.1.2 5.522434e-06 0.3569886 0.009 0.047 2.021321e-01 2
## GKAP1.2 5.529283e-06 -0.3309631 0.012 0.053 2.023828e-01 2
## USPL1 5.560226e-06 0.9542810 0.057 0.124 2.035154e-01 2
## AC010894.3.1 5.578755e-06 0.6358889 0.051 0.115 2.041936e-01 2
## LIG4 5.620524e-06 -0.1594585 0.012 0.053 2.057224e-01 2
## KPNA6.1 5.641252e-06 0.8589016 0.072 0.147 2.064811e-01 2
## ZGRF1.2 5.657042e-06 0.6498303 0.018 0.063 2.070590e-01 2
## POLA1.2 5.674341e-06 0.8123122 0.057 0.123 2.076922e-01 2
## ZNF569 5.682715e-06 0.2728125 0.018 0.063 2.079987e-01 2
## WDR33.1 5.702487e-06 1.0348886 0.189 0.337 2.087224e-01 2
## ZNF586 5.769358e-06 0.6137489 0.026 0.075 2.111701e-01 2
## PSTK 5.771093e-06 -1.3847329 0.003 0.036 2.112336e-01 2
## ZNF836 5.807704e-06 -0.3754520 0.015 0.057 2.125736e-01 2
## AC016876.1 5.812540e-06 -0.7039268 0.005 0.039 2.127506e-01 2
## FMR1 5.813203e-06 0.7635694 0.071 0.144 2.127749e-01 2
## JADE2 5.892319e-06 0.7339201 0.066 0.137 2.156707e-01 2
## CUL4B 5.897451e-06 0.7878427 0.051 0.114 2.158585e-01 2
## ZC4H2 5.954722e-06 0.6895238 0.056 0.121 2.179547e-01 2
## GPATCH3 5.988438e-06 0.5554419 0.026 0.075 2.191888e-01 2
## ARHGAP45.1 6.014706e-06 0.9203817 0.272 0.485 2.201503e-01 2
## LXN.1 6.018541e-06 0.1354638 0.015 0.058 2.202906e-01 2
## PPIL2.1 6.041741e-06 1.0473077 0.074 0.149 2.211398e-01 2
## USP33 6.118458e-06 0.9913392 0.111 0.207 2.239478e-01 2
## TBC1D1.1 6.120651e-06 0.9596507 0.138 0.249 2.240281e-01 2
## FXYD2.1 6.121896e-06 -1.7656387 0.005 0.039 2.240737e-01 2
## DHX58 6.138365e-06 0.4078230 0.012 0.052 2.246764e-01 2
## ZG16B 6.177860e-06 -1.7451280 0.003 0.036 2.261220e-01 2
## ZBTB2.1 6.198782e-06 0.7878800 0.045 0.105 2.268878e-01 2
## NCALD.1 6.209651e-06 2.1588754 0.287 0.274 2.272857e-01 2
## IFNGR1.2 6.211909e-06 0.6380942 0.093 0.177 2.273683e-01 2
## KCTD9.1 6.249597e-06 0.6251536 0.042 0.100 2.287478e-01 2
## AL135818.1.2 6.270323e-06 1.1483520 0.057 0.123 2.295064e-01 2
## SPTBN1.2 6.272534e-06 0.9507462 0.212 0.372 2.295873e-01 2
## PDIK1L.1 6.282396e-06 0.5934269 0.020 0.065 2.299483e-01 2
## IRAK1BP1 6.303233e-06 0.5782166 0.027 0.077 2.307109e-01 2
## SLC27A2.1 6.303328e-06 0.3480659 0.020 0.065 2.307144e-01 2
## C1orf112.2 6.309107e-06 0.3163345 0.024 0.072 2.309259e-01 2
## RAD17 6.311798e-06 0.7286112 0.048 0.110 2.310244e-01 2
## NAA15.1 6.314141e-06 1.1691998 0.110 0.204 2.311102e-01 2
## ZNF789.1 6.316838e-06 0.6237219 0.035 0.088 2.312089e-01 2
## TPP2.1 6.327482e-06 0.9330116 0.192 0.339 2.315985e-01 2
## CEP83.1 6.405108e-06 1.0460601 0.060 0.128 2.344398e-01 2
## AAK1 6.415672e-06 1.8684423 0.322 0.327 2.348264e-01 2
## GEMIN5 6.419133e-06 0.3716631 0.026 0.074 2.349531e-01 2
## TRIM13 6.505921e-06 0.8795766 0.047 0.107 2.381297e-01 2
## QTRT2 6.579782e-06 0.5199508 0.035 0.088 2.408332e-01 2
## ZNF257.2 6.621667e-06 -0.4254966 0.009 0.047 2.423663e-01 2
## PPP2R1B.2 6.622898e-06 0.5794053 0.041 0.098 2.424113e-01 2
## CLASRP 6.639101e-06 1.1817587 0.075 0.151 2.430044e-01 2
## TBC1D22A 6.649132e-06 1.8704646 0.344 0.366 2.433715e-01 2
## SREK1 6.659469e-06 0.9739205 0.164 0.291 2.437499e-01 2
## LLGL1 6.675279e-06 -1.4509869 0.005 0.039 2.443286e-01 2
## PIP4K2C 6.712933e-06 0.5910161 0.023 0.070 2.457068e-01 2
## EXOC1 6.748528e-06 1.0627752 0.084 0.165 2.470096e-01 2
## FXYD7.1 6.788170e-06 -2.1015877 0.002 0.033 2.484606e-01 2
## IDNK 6.803196e-06 0.7254492 0.027 0.077 2.490106e-01 2
## TTC31 6.813055e-06 0.4677994 0.029 0.079 2.493714e-01 2
## INSIG1 6.824004e-06 1.1191127 0.077 0.153 2.497722e-01 2
## TRBV7-2 6.853437e-06 -1.8147863 0.027 0.074 2.508495e-01 2
## ZNF592 6.887629e-06 0.9202893 0.066 0.137 2.521010e-01 2
## HINT3.1 6.901611e-06 0.5633043 0.015 0.057 2.526128e-01 2
## PAXIP1-AS2 6.909708e-06 0.1658347 0.030 0.081 2.529091e-01 2
## PNPLA6 6.913953e-06 1.1539013 0.050 0.111 2.530645e-01 2
## CPT1A.2 6.928434e-06 0.9717761 0.123 0.225 2.535945e-01 2
## CASTOR2 6.971848e-06 0.7948398 0.045 0.104 2.551836e-01 2
## NFE2L2 6.978306e-06 1.0452703 0.165 0.294 2.554200e-01 2
## RMND1 6.982455e-06 0.9060255 0.042 0.100 2.555718e-01 2
## STAG2.1 6.988717e-06 0.9446256 0.209 0.371 2.558010e-01 2
## PTCD1 7.025326e-06 0.4005599 0.014 0.055 2.571410e-01 2
## LIN7C 7.049000e-06 0.5745953 0.039 0.095 2.580075e-01 2
## ZDHHC5.1 7.074528e-06 0.9639312 0.065 0.135 2.589419e-01 2
## SACM1L.1 7.149672e-06 1.2207242 0.168 0.299 2.616923e-01 2
## ABCB10.1 7.162086e-06 0.8452192 0.054 0.118 2.621467e-01 2
## ENDOV 7.257548e-06 0.9221049 0.021 0.067 2.656408e-01 2
## VPS11 7.274019e-06 0.7626493 0.038 0.093 2.662436e-01 2
## CDC25C.2 7.277684e-06 -1.2993726 0.002 0.033 2.663778e-01 2
## RBFOX2 7.280512e-06 -1.0545361 0.002 0.033 2.664813e-01 2
## POMT1.1 7.285021e-06 0.9462645 0.036 0.091 2.666463e-01 2
## SLC35A2 7.306014e-06 0.5167650 0.018 0.062 2.674147e-01 2
## EMC1 7.306271e-06 0.4433998 0.036 0.090 2.674241e-01 2
## RNF121.2 7.329029e-06 0.6917373 0.042 0.100 2.682571e-01 2
## MFSD14A 7.403272e-06 1.0418791 0.146 0.262 2.709746e-01 2
## ZFYVE19.1 7.518875e-06 0.7584664 0.029 0.079 2.752058e-01 2
## AFMID.2 7.599630e-06 0.6775412 0.011 0.049 2.781617e-01 2
## CHUK 7.605804e-06 0.8980988 0.039 0.095 2.783877e-01 2
## SNHG10 7.649145e-06 -0.8520488 0.005 0.038 2.799740e-01 2
## SAMD9L.1 7.661120e-06 0.9970174 0.126 0.229 2.804123e-01 2
## ATP8B2.1 7.698226e-06 0.9227913 0.150 0.268 2.817705e-01 2
## OXR1 7.729222e-06 0.6314107 0.059 0.125 2.829050e-01 2
## ZNF174 7.742896e-06 0.3803164 0.015 0.057 2.834055e-01 2
## ZMYM1 7.771553e-06 0.2491162 0.023 0.069 2.844544e-01 2
## MBNL3.1 7.772841e-06 0.4744618 0.045 0.104 2.845015e-01 2
## B4GALT2 7.802444e-06 -2.1383950 0.002 0.032 2.855851e-01 2
## ANKRD44-IT1 7.802486e-06 3.6877824 0.018 0.004 2.855866e-01 2
## TRAV8-6 7.803671e-06 -0.5174233 0.011 0.049 2.856300e-01 2
## YTHDC1.1 7.913797e-06 1.1329515 0.183 0.326 2.896608e-01 2
## GPR68.1 7.925834e-06 1.2303823 0.111 0.206 2.901014e-01 2
## CRELD1 7.933439e-06 0.5913500 0.030 0.081 2.903797e-01 2
## STAT5B 8.007986e-06 0.8933253 0.174 0.306 2.931083e-01 2
## COLGALT1.1 8.049395e-06 0.6924758 0.023 0.069 2.946240e-01 2
## ARRDC1-AS1 8.059735e-06 0.7281911 0.021 0.067 2.950024e-01 2
## CEP152.2 8.062618e-06 0.7449745 0.044 0.102 2.951079e-01 2
## STAT3 8.066107e-06 1.0500802 0.211 0.366 2.952357e-01 2
## ABHD11.1 8.098394e-06 0.4052886 0.017 0.059 2.964174e-01 2
## TOMM40L.2 8.099054e-06 -0.1554898 0.011 0.049 2.964416e-01 2
## CEP72.2 8.099312e-06 -1.7301843 0.005 0.038 2.964510e-01 2
## RPRD2 8.101625e-06 0.9655089 0.132 0.239 2.965357e-01 2
## FAM3C.1 8.129376e-06 0.6632592 0.069 0.140 2.975514e-01 2
## WASHC4 8.157917e-06 0.8286677 0.126 0.227 2.985961e-01 2
## LINC02084.1 8.168884e-06 -5.4380980 0.000 0.029 2.989975e-01 2
## PGPEP1 8.169743e-06 1.1150104 0.032 0.083 2.990289e-01 2
## PUS3 8.208294e-06 0.8364768 0.014 0.054 3.004400e-01 2
## CEP85.2 8.217689e-06 0.6335089 0.024 0.071 3.007839e-01 2
## AC245014.3.1 8.324835e-06 0.7345667 0.108 0.199 3.047056e-01 2
## TRBV9.2 8.473197e-06 -1.5617021 0.023 0.067 3.101359e-01 2
## NFIC.1 8.512134e-06 0.6789496 0.045 0.104 3.115611e-01 2
## ZBTB38.1 8.529292e-06 0.8991859 0.126 0.227 3.121891e-01 2
## SPRTN 8.536234e-06 0.9281320 0.048 0.108 3.124433e-01 2
## GPN2.1 8.539194e-06 1.1525390 0.033 0.085 3.125516e-01 2
## RANBP9.1 8.584331e-06 1.1604829 0.126 0.229 3.142037e-01 2
## TGFBR3L 8.606226e-06 -0.6007120 0.009 0.046 3.150051e-01 2
## ANAPC4 8.660569e-06 0.8879284 0.075 0.149 3.169941e-01 2
## ADGRE1.2 8.708801e-06 -1.5739350 0.003 0.035 3.187595e-01 2
## TRAPPC2 8.710098e-06 0.7687026 0.042 0.099 3.188070e-01 2
## CHPF2 8.744856e-06 0.4248041 0.026 0.073 3.200792e-01 2
## ESPL1.2 8.759457e-06 -5.2549520 0.000 0.029 3.206136e-01 2
## FANCL.2 8.772256e-06 0.5342715 0.024 0.071 3.210821e-01 2
## ZSCAN21 8.780174e-06 -0.9333933 0.005 0.038 3.213719e-01 2
## EZH2.2 8.781003e-06 0.7218178 0.144 0.254 3.214023e-01 2
## NSD2.2 8.867369e-06 0.9043177 0.152 0.267 3.245635e-01 2
## AKAP1.1 8.886313e-06 0.8451000 0.024 0.071 3.252568e-01 2
## ARMC9 8.968023e-06 -2.5137100 0.002 0.032 3.282476e-01 2
## EXOG 8.975983e-06 1.0516916 0.042 0.099 3.285389e-01 2
## SENP6 9.006987e-06 0.8840073 0.220 0.384 3.296737e-01 2
## PRDM4.1 9.021165e-06 0.7171672 0.032 0.083 3.301927e-01 2
## RNPC3 9.029203e-06 1.2617618 0.080 0.157 3.304869e-01 2
## SMPD2 9.044194e-06 0.6088724 0.026 0.073 3.310356e-01 2
## ENDOD1.1 9.119047e-06 0.8161256 0.035 0.087 3.337754e-01 2
## RETREG3 9.165675e-06 0.8771767 0.047 0.106 3.354820e-01 2
## HSF2.1 9.201051e-06 0.6210524 0.026 0.073 3.367769e-01 2
## TRMU.1 9.237130e-06 1.1195708 0.044 0.101 3.380974e-01 2
## GLA.1 9.254751e-06 0.6959805 0.053 0.115 3.387424e-01 2
## SPTLC2.2 9.286010e-06 1.1871000 0.108 0.200 3.398865e-01 2
## CBR4 9.305432e-06 0.6939934 0.027 0.076 3.405974e-01 2
## CBY1 9.442311e-06 0.1068922 0.017 0.059 3.456075e-01 2
## IQCG 9.466385e-06 0.6227439 0.023 0.068 3.464886e-01 2
## VNN2.1 9.591720e-06 0.6675889 0.026 0.073 3.510761e-01 2
## PRKCQ.1 9.614594e-06 1.8453784 0.364 0.402 3.519134e-01 2
## TRAV35 9.618206e-06 -2.2833420 0.002 0.032 3.520456e-01 2
## PAK1 9.790620e-06 0.7341143 0.045 0.103 3.583563e-01 2
## RSF1.1 9.801727e-06 0.8690663 0.236 0.414 3.587628e-01 2
## FBXO28 9.857899e-06 0.7966120 0.050 0.110 3.608188e-01 2
## TRAV4 9.876099e-06 -1.9319172 0.006 0.040 3.614850e-01 2
## ZNF672 9.972058e-06 0.5048276 0.027 0.075 3.649973e-01 2
## PPP3CC.2 1.001799e-05 1.0859536 0.195 0.342 3.666783e-01 2
## SESN3.2 1.002598e-05 0.9153886 0.114 0.208 3.669711e-01 2
## LIN52.2 1.009157e-05 1.0423492 0.054 0.117 3.693716e-01 2
## CCDC174 1.020226e-05 1.0633900 0.080 0.156 3.734231e-01 2
## FAM53C 1.026286e-05 0.7400130 0.041 0.096 3.756411e-01 2
## NEIL2.1 1.031146e-05 -1.4371020 0.002 0.032 3.774200e-01 2
## NUDT7 1.031146e-05 -1.6804547 0.002 0.032 3.774200e-01 2
## CREB1 1.040692e-05 1.0454418 0.162 0.287 3.809140e-01 2
## ZNF250 1.044937e-05 0.6174913 0.021 0.066 3.824679e-01 2
## NCDN.1 1.052551e-05 0.5384765 0.021 0.066 3.852546e-01 2
## SMG8 1.054795e-05 0.1031935 0.014 0.053 3.860761e-01 2
## ZNF41.1 1.056094e-05 0.3757118 0.023 0.068 3.865516e-01 2
## BTD 1.056371e-05 0.4162977 0.042 0.098 3.866530e-01 2
## GADD45G.2 1.058834e-05 0.9790267 0.116 0.208 3.875543e-01 2
## PARP6 1.060219e-05 0.6990301 0.042 0.099 3.880615e-01 2
## ASXL1.1 1.061371e-05 1.1272592 0.186 0.327 3.884829e-01 2
## CAPN7.1 1.063072e-05 1.1382513 0.113 0.207 3.891056e-01 2
## ZNF281 1.068492e-05 0.2758208 0.041 0.096 3.910893e-01 2
## NARS2.1 1.074242e-05 1.0011369 0.053 0.114 3.931942e-01 2
## SERPINH1.1 1.076249e-05 -0.1785975 0.008 0.043 3.939286e-01 2
## TRBV2.2 1.087590e-05 -2.4883368 0.011 0.047 3.980799e-01 2
## AC127521.1.1 1.091270e-05 0.3260930 0.024 0.070 3.994265e-01 2
## STXBP1 1.100959e-05 0.2551278 0.020 0.063 4.029731e-01 2
## PRKRIP1 1.102492e-05 1.2151502 0.119 0.217 4.035339e-01 2
## MXI1 1.104036e-05 1.2724649 0.069 0.140 4.040993e-01 2
## STOML1.1 1.104604e-05 -2.3749928 0.002 0.031 4.043070e-01 2
## TRAV27.1 1.111687e-05 -1.3882531 0.012 0.050 4.068995e-01 2
## GOLGA8A.1 1.112370e-05 2.6150610 0.194 0.155 4.071495e-01 2
## TTC14 1.113604e-05 1.0057301 0.129 0.233 4.076014e-01 2
## DDX59 1.117573e-05 0.9923159 0.048 0.107 4.090541e-01 2
## IL15RA.2 1.122188e-05 0.8102975 0.044 0.101 4.107432e-01 2
## FEM1A 1.124979e-05 0.4073896 0.018 0.061 4.117648e-01 2
## TOR1B 1.127570e-05 0.7302615 0.036 0.089 4.127131e-01 2
## KIAA1958 1.129988e-05 -0.5723853 0.009 0.045 4.135983e-01 2
## SLC25A40.1 1.132100e-05 0.6442906 0.056 0.118 4.143712e-01 2
## ZNF75A 1.135020e-05 0.7626896 0.047 0.105 4.154400e-01 2
## ITK.1 1.137349e-05 1.4942539 0.426 0.522 4.162925e-01 2
## ASB16-AS1 1.140081e-05 1.0870740 0.027 0.075 4.172924e-01 2
## C5orf63 1.151325e-05 -1.3824532 0.003 0.034 4.214079e-01 2
## C11orf98 1.151751e-05 -1.4541597 0.003 0.034 4.215640e-01 2
## BOD1L1.1 1.152498e-05 0.9380259 0.224 0.393 4.218372e-01 2
## DDX60 1.161579e-05 0.8394625 0.051 0.112 4.251610e-01 2
## CEP120 1.167985e-05 1.0965103 0.084 0.162 4.275060e-01 2
## SMKR1 1.184206e-05 -2.2265042 0.002 0.031 4.334432e-01 2
## CENPA.2 1.210136e-05 0.4104425 0.011 0.048 4.429341e-01 2
## FBLN7.2 1.214246e-05 -0.6474968 0.008 0.043 4.444383e-01 2
## TMEM44-AS1 1.215250e-05 0.6222617 0.018 0.060 4.448058e-01 2
## TRIM14.1 1.218410e-05 0.9337926 0.098 0.183 4.459626e-01 2
## CHEK2.2 1.223938e-05 0.7918178 0.021 0.065 4.479858e-01 2
## PCNX1.1 1.231652e-05 1.7692729 0.373 0.423 4.508091e-01 2
## TMEM185B 1.234944e-05 -0.2803271 0.021 0.065 4.520143e-01 2
## PHC1 1.254173e-05 0.5005616 0.027 0.075 4.590525e-01 2
## ZNF567 1.257331e-05 0.7518402 0.042 0.098 4.602084e-01 2
## MED9 1.267136e-05 0.8425577 0.021 0.065 4.637971e-01 2
## NDE1 1.277736e-05 1.2047711 0.132 0.236 4.676769e-01 2
## CEP57L1.1 1.278465e-05 0.7773045 0.030 0.079 4.679439e-01 2
## PCMTD1 1.290635e-05 1.0072683 0.156 0.276 4.723980e-01 2
## COG8 1.294383e-05 0.5795683 0.033 0.084 4.737699e-01 2
## AVEN 1.296262e-05 1.1246359 0.050 0.109 4.744577e-01 2
## LRRFIP2.1 1.301268e-05 1.1999416 0.114 0.208 4.762900e-01 2
## GNG2.1 1.304294e-05 1.1959909 0.508 0.684 4.773979e-01 2
## EXT2 1.307317e-05 1.1200743 0.057 0.121 4.785042e-01 2
## XPNPEP3 1.318215e-05 1.0579761 0.039 0.093 4.824930e-01 2
## BIRC6 1.320781e-05 1.5569437 0.395 0.466 4.834322e-01 2
## MTHFD1L.2 1.322357e-05 1.0081075 0.044 0.100 4.840091e-01 2
## FZR1.1 1.323231e-05 0.7445991 0.035 0.086 4.843289e-01 2
## ZNF3 1.329989e-05 0.4749325 0.026 0.072 4.868026e-01 2
## CD302.1 1.331804e-05 0.5256948 0.024 0.070 4.874668e-01 2
## ZNF487 1.340404e-05 0.8233111 0.021 0.065 4.906148e-01 2
## MANEA-DT 1.352019e-05 0.5341807 0.023 0.067 4.948659e-01 2
## SCARB2.1 1.352224e-05 -0.2438825 0.014 0.053 4.949409e-01 2
## HASPIN.2 1.362068e-05 -0.5160953 0.002 0.031 4.985440e-01 2
## MTMR6 1.363435e-05 0.9825563 0.095 0.177 4.990446e-01 2
## PEX6 1.367456e-05 0.9478116 0.032 0.082 5.005162e-01 2
## FAM111A-DT 1.374271e-05 -0.9997649 0.006 0.039 5.030106e-01 2
## AKAP8 1.382538e-05 0.9541696 0.074 0.145 5.060366e-01 2
## SAMD4B 1.382952e-05 0.9127217 0.045 0.102 5.061879e-01 2
## CBX6.1 1.391907e-05 0.8613031 0.057 0.120 5.094656e-01 2
## SRF 1.398756e-05 0.9070547 0.047 0.104 5.119726e-01 2
## TRAV26-1 1.403007e-05 -0.9355796 0.017 0.057 5.135287e-01 2
## TENT4B 1.411618e-05 0.9768153 0.041 0.095 5.166805e-01 2
## KCTD13 1.411690e-05 0.5807156 0.045 0.102 5.167069e-01 2
## HS1BP3 1.412470e-05 0.7614523 0.035 0.086 5.169922e-01 2
## HACL1 1.417471e-05 1.0293034 0.066 0.134 5.188228e-01 2
## NLRX1.1 1.420889e-05 -0.8361312 0.011 0.047 5.200737e-01 2
## MECOM 1.427685e-05 -5.0043145 0.000 0.028 5.225613e-01 2
## AL662797.1.2 1.427685e-05 -4.4943113 0.000 0.028 5.225613e-01 2
## RBM28.1 1.432407e-05 0.9943699 0.062 0.127 5.242896e-01 2
## MINDY2 1.438553e-05 0.8614783 0.080 0.154 5.265393e-01 2
## AURKA.2 1.440858e-05 0.8968444 0.014 0.052 5.273828e-01 2
## GEN1.2 1.450810e-05 0.5842937 0.029 0.076 5.310255e-01 2
## CASC4 1.451424e-05 1.0756164 0.134 0.238 5.312504e-01 2
## FTX 1.453189e-05 2.1925593 0.278 0.267 5.318962e-01 2
## CXorf40A 1.457353e-05 0.7545785 0.051 0.111 5.334202e-01 2
## MFSD3 1.461327e-05 -0.2151452 0.015 0.055 5.348748e-01 2
## TRAPPC12 1.465684e-05 0.9944474 0.102 0.189 5.364698e-01 2
## CLCC1.1 1.477681e-05 0.6224043 0.015 0.055 5.408609e-01 2
## TBC1D9B.1 1.481102e-05 1.2197774 0.066 0.134 5.421131e-01 2
## MARF1 1.481630e-05 0.9146705 0.084 0.161 5.423063e-01 2
## PEX10 1.495210e-05 0.5855988 0.018 0.060 5.472766e-01 2
## ZFP36.2 1.503216e-05 0.4897611 0.388 0.658 5.502070e-01 2
## NDST2 1.503770e-05 0.9275225 0.038 0.091 5.504098e-01 2
## PCMTD2 1.513149e-05 0.7054484 0.059 0.122 5.538429e-01 2
## CDK11A 1.521573e-05 1.1871716 0.105 0.194 5.569261e-01 2
## PARP2.1 1.534629e-05 0.7824709 0.023 0.067 5.617050e-01 2
## PKIA.1 1.536517e-05 0.3984164 0.021 0.064 5.623960e-01 2
## MEX3C 1.539229e-05 1.0074189 0.047 0.104 5.633887e-01 2
## BCL9L.2 1.541899e-05 0.6559100 0.056 0.118 5.643660e-01 2
## TRIM24.2 1.549049e-05 1.0353292 0.068 0.136 5.669831e-01 2
## AP003086.1.1 1.550056e-05 1.1245783 0.074 0.145 5.673517e-01 2
## DIPK2A 1.550750e-05 1.0444749 0.056 0.118 5.676054e-01 2
## OSM.2 1.554964e-05 0.1742419 0.036 0.087 5.691478e-01 2
## PLEKHA1.2 1.564764e-05 0.6454466 0.050 0.109 5.727349e-01 2
## PDSS1.2 1.586360e-05 0.9328280 0.024 0.069 5.806395e-01 2
## SH3BP2.1 1.600355e-05 0.9056553 0.060 0.124 5.857619e-01 2
## ATF5.1 1.606275e-05 0.7705362 0.038 0.090 5.879286e-01 2
## VIPAS39 1.631921e-05 0.5559286 0.029 0.076 5.973156e-01 2
## KRT1.2 1.641484e-05 -5.3409683 0.000 0.027 6.008161e-01 2
## MTMR2.1 1.641679e-05 0.7266976 0.042 0.097 6.008872e-01 2
## SETDB1 1.644306e-05 0.9619704 0.051 0.111 6.018488e-01 2
## WDR11 1.644787e-05 0.9878209 0.072 0.142 6.020251e-01 2
## ZC3H3.1 1.661352e-05 1.1128513 0.057 0.120 6.080879e-01 2
## LINC02245.1 1.676342e-05 1.0476487 0.042 0.097 6.135746e-01 2
## IFT74.1 1.685673e-05 0.2787160 0.026 0.071 6.169901e-01 2
## HMGXB3 1.686837e-05 1.0442521 0.090 0.170 6.174159e-01 2
## CHI3L2.2 1.689733e-05 -0.2127813 0.006 0.039 6.184759e-01 2
## CCDC93.1 1.698537e-05 0.9659294 0.069 0.137 6.216984e-01 2
## P2RX5.2 1.704730e-05 0.7934139 0.033 0.083 6.239652e-01 2
## SUPT5H 1.714105e-05 1.1621543 0.150 0.264 6.273966e-01 2
## DYNLRB1 1.729421e-05 0.2602212 0.014 0.052 6.330027e-01 2
## SREBF1.2 1.740576e-05 0.8976185 0.039 0.092 6.370857e-01 2
## TLR9 1.741430e-05 -1.9466414 0.003 0.033 6.373983e-01 2
## KIF1C.1 1.749855e-05 0.7829357 0.027 0.074 6.404819e-01 2
## UBE4B.1 1.752557e-05 1.1497404 0.098 0.181 6.414711e-01 2
## SLC17A5 1.754310e-05 0.4216482 0.035 0.085 6.421127e-01 2
## TRAPPC11 1.771561e-05 0.6797209 0.056 0.117 6.484267e-01 2
## DHX33.2 1.774693e-05 0.4540339 0.023 0.067 6.495731e-01 2
## RENBP 1.792209e-05 0.4254643 0.020 0.062 6.559845e-01 2
## UBXN2B 1.800869e-05 0.5983771 0.029 0.076 6.591539e-01 2
## TMEM41B 1.801423e-05 1.0770307 0.077 0.149 6.593569e-01 2
## TRMT1L 1.809609e-05 0.9459894 0.054 0.115 6.623530e-01 2
## TRAV23DV6 1.814430e-05 -1.8540279 0.009 0.044 6.641179e-01 2
## UGCG 1.817815e-05 0.9117250 0.059 0.122 6.653565e-01 2
## CAPN12.2 1.824772e-05 0.7279982 0.057 0.119 6.679032e-01 2
## ITGA5 1.828436e-05 0.6362664 0.030 0.078 6.692443e-01 2
## FOXP3 1.831680e-05 -1.7706114 0.005 0.036 6.704316e-01 2
## EFR3A 1.848094e-05 0.9403076 0.159 0.279 6.764394e-01 2
## MPZL3.1 1.854964e-05 1.0828872 0.104 0.190 6.789538e-01 2
## TMEM71.1 1.866016e-05 1.0596335 0.116 0.209 6.829993e-01 2
## ZNF747 1.867309e-05 -1.6408384 0.003 0.033 6.834726e-01 2
## SFSWAP 1.904570e-05 1.0261416 0.141 0.249 6.971107e-01 2
## PRKCA-AS1.1 1.906774e-05 0.8499887 0.036 0.087 6.979176e-01 2
## ZNF213-AS1 1.911002e-05 0.4456943 0.027 0.073 6.994650e-01 2
## ADAT2 1.927867e-05 0.4827714 0.021 0.064 7.056380e-01 2
## WWOX.2 1.932281e-05 1.9609011 0.316 0.318 7.072536e-01 2
## NORAD 1.943824e-05 0.8560909 0.165 0.286 7.114783e-01 2
## EDC4.1 1.944841e-05 1.1245005 0.053 0.112 7.118506e-01 2
## SCAMP1-AS1 1.947559e-05 0.7736679 0.020 0.061 7.128454e-01 2
## ZNF362 1.948720e-05 0.5833739 0.032 0.080 7.132706e-01 2
## ZNF579 1.956298e-05 -0.3100686 0.008 0.041 7.160441e-01 2
## AC009779.2 1.964140e-05 -1.2492408 0.005 0.036 7.189144e-01 2
## TCEANC2.2 1.965868e-05 0.7245632 0.023 0.066 7.195472e-01 2
## ZNF568 1.982137e-05 -0.2704297 0.011 0.046 7.255016e-01 2
## FAM174A 1.982824e-05 0.4519390 0.024 0.068 7.257533e-01 2
## IGHMBP2 1.992265e-05 0.3752223 0.021 0.064 7.292088e-01 2
## GIGYF2.1 1.992483e-05 1.1781477 0.152 0.267 7.292885e-01 2
## PPRC1.1 1.992499e-05 0.9570235 0.042 0.096 7.292946e-01 2
## MED27.1 2.010589e-05 1.0700113 0.119 0.214 7.359159e-01 2
## ACSS1.1 2.022703e-05 0.9089070 0.062 0.126 7.403499e-01 2
## TMEM63A.2 2.029917e-05 0.8553792 0.075 0.146 7.429901e-01 2
## SIRT3 2.031467e-05 0.8551053 0.029 0.075 7.435576e-01 2
## FAM227B 2.060767e-05 0.2468504 0.015 0.054 7.542820e-01 2
## AXIN1.1 2.061035e-05 0.7019680 0.057 0.118 7.543801e-01 2
## ADAMTS6 2.062604e-05 0.2986403 0.020 0.061 7.549543e-01 2
## CHD1 2.064941e-05 1.0165257 0.215 0.374 7.558096e-01 2
## IER3.1 2.066337e-05 -0.1057683 0.008 0.041 7.563206e-01 2
## GTF2E1 2.066695e-05 0.1430951 0.015 0.054 7.564517e-01 2
## AC005562.1 2.075720e-05 -0.4391587 0.009 0.044 7.597551e-01 2
## ZNF197 2.082342e-05 -0.5042603 0.009 0.044 7.621789e-01 2
## AC092652.2 2.105388e-05 0.4811050 0.026 0.071 7.706140e-01 2
## MIGA2 2.107517e-05 0.8605553 0.030 0.078 7.713932e-01 2
## SLAIN1 2.110822e-05 0.6815979 0.027 0.073 7.726029e-01 2
## CDK16 2.112321e-05 1.0506374 0.045 0.101 7.731519e-01 2
## CREB3L2.1 2.124241e-05 1.0376100 0.089 0.166 7.775146e-01 2
## GRAP.1 2.135719e-05 0.7373510 0.014 0.051 7.817158e-01 2
## CTNS 2.136973e-05 0.6233256 0.014 0.051 7.821749e-01 2
## AP001347.1.2 2.138172e-05 0.2501407 0.018 0.059 7.826136e-01 2
## ITPRID2.1 2.142637e-05 0.5691460 0.032 0.080 7.842480e-01 2
## THAP7-AS1.1 2.143001e-05 -0.8405992 0.003 0.033 7.843812e-01 2
## INPP5D.1 2.162980e-05 1.8153401 0.350 0.384 7.916940e-01 2
## TDP1.1 2.165154e-05 1.0032342 0.074 0.144 7.924895e-01 2
## LY6G5C 2.169881e-05 -4.9102475 0.000 0.026 7.942199e-01 2
## MSANTD4 2.189210e-05 -0.1954931 0.014 0.051 8.012947e-01 2
## HIBCH 2.195959e-05 1.1705302 0.087 0.164 8.037648e-01 2
## SLC9B2 2.200210e-05 0.6685020 0.044 0.098 8.053208e-01 2
## AL050341.2 2.203693e-05 -0.7583983 0.006 0.038 8.065957e-01 2
## CDKN1A.1 2.207940e-05 0.9205714 0.089 0.164 8.081502e-01 2
## MORN2.1 2.215561e-05 -1.7415681 0.002 0.030 8.109397e-01 2
## C19orf54 2.219643e-05 0.2876426 0.009 0.043 8.124338e-01 2
## D2HGDH 2.223037e-05 0.5264551 0.027 0.073 8.136760e-01 2
## TMEM38B.2 2.228493e-05 0.8236030 0.024 0.068 8.156729e-01 2
## COP1.1 2.237842e-05 1.7872711 0.344 0.374 8.190950e-01 2
## ADPRM 2.245030e-05 0.4112896 0.024 0.068 8.217258e-01 2
## MTBP.2 2.277626e-05 0.7426553 0.015 0.054 8.336568e-01 2
## SLC46A3 2.304183e-05 0.3647132 0.020 0.061 8.433771e-01 2
## CTDSPL2.1 2.328537e-05 1.0108294 0.080 0.152 8.522911e-01 2
## TFDP2.1 2.344801e-05 1.1566827 0.086 0.161 8.582442e-01 2
## ST3GAL3.1 2.349709e-05 0.8881003 0.063 0.127 8.600405e-01 2
## AC023424.3 2.353294e-05 0.8705986 0.056 0.116 8.613525e-01 2
## ZNF600 2.363326e-05 0.4987988 0.027 0.073 8.650248e-01 2
## KPNA5 2.364109e-05 0.3733129 0.041 0.093 8.653112e-01 2
## ALG10 2.377523e-05 -0.6076456 0.008 0.041 8.702209e-01 2
## PIGQ 2.380511e-05 0.6165833 0.021 0.063 8.713148e-01 2
## SGK3 2.382198e-05 1.1416922 0.069 0.136 8.719322e-01 2
## TNFAIP1 2.395700e-05 0.3692985 0.015 0.053 8.768740e-01 2
## LTN1 2.397352e-05 1.0589790 0.105 0.191 8.774786e-01 2
## RAP2C 2.404799e-05 0.6222788 0.041 0.093 8.802044e-01 2
## ZNF350 2.404852e-05 0.2673809 0.020 0.061 8.802241e-01 2
## VIRMA 2.413668e-05 1.0487781 0.102 0.186 8.834508e-01 2
## HERC6 2.420872e-05 0.8180278 0.036 0.086 8.860874e-01 2
## DHX38 2.425778e-05 1.1912464 0.069 0.136 8.878833e-01 2
## CCDC125 2.439251e-05 0.7048299 0.021 0.063 8.928148e-01 2
## AC244090.1.1 2.457707e-05 -0.7743022 0.005 0.035 8.995699e-01 2
## NXPE3 2.464166e-05 0.5818628 0.047 0.102 9.019340e-01 2
## DNAJC14 2.474424e-05 0.4321522 0.024 0.067 9.056885e-01 2
## UNC13D.1 2.482024e-05 1.2857356 0.117 0.210 9.084705e-01 2
## SPACA9 2.494784e-05 -4.9012642 0.000 0.026 9.131407e-01 2
## ICE2.2 2.504628e-05 1.1693408 0.111 0.201 9.167441e-01 2
## TMEM177 2.511402e-05 -0.3933526 0.009 0.043 9.192235e-01 2
## LRFN4 2.511992e-05 0.4706385 0.017 0.056 9.194393e-01 2
## ZNF518A 2.513424e-05 0.6495353 0.054 0.113 9.199636e-01 2
## ZBTB25 2.513694e-05 1.0751470 0.128 0.227 9.200624e-01 2
## TSPOAP1-AS1 2.518532e-05 1.1213206 0.129 0.228 9.218329e-01 2
## FGFBP2.2 2.529548e-05 -2.5298890 0.005 0.035 9.258650e-01 2
## SHISAL2A 2.534017e-05 0.5801827 0.023 0.065 9.275009e-01 2
## NDUFAF7 2.540732e-05 1.0493861 0.036 0.086 9.299589e-01 2
## VAV1 2.547212e-05 1.1820396 0.135 0.238 9.323305e-01 2
## FAM234A.1 2.557327e-05 0.3740413 0.032 0.079 9.360329e-01 2
## KIZ 2.561301e-05 0.9670514 0.059 0.120 9.374873e-01 2
## SLC5A6 2.562926e-05 0.6732694 0.024 0.068 9.380820e-01 2
## ZSWIM9 2.564887e-05 0.8556865 0.015 0.053 9.388000e-01 2
## ELP1 2.569107e-05 0.8464242 0.056 0.116 9.403445e-01 2
## LRRC57.1 2.571175e-05 0.6358658 0.021 0.063 9.411015e-01 2
## ARID1A.1 2.572011e-05 1.0644950 0.165 0.287 9.414076e-01 2
## CPOX.1 2.573488e-05 0.8794720 0.060 0.122 9.419481e-01 2
## TAF3 2.577798e-05 1.0828582 0.149 0.260 9.435255e-01 2
## YME1L1 2.609782e-05 1.1969883 0.281 0.491 9.552325e-01 2
## SIRT6 2.616303e-05 0.9403128 0.026 0.070 9.576191e-01 2
## PLEKHG3.1 2.631183e-05 0.4678818 0.027 0.072 9.630657e-01 2
## YWHAG.2 2.659327e-05 1.0721395 0.099 0.181 9.733669e-01 2
## BANP 2.665234e-05 1.0886384 0.069 0.136 9.755288e-01 2
## PIK3R4 2.676900e-05 0.8703537 0.060 0.122 9.797990e-01 2
## ZMYM4.1 2.677796e-05 1.2170031 0.104 0.189 9.801268e-01 2
## JPX 2.727775e-05 1.1597477 0.120 0.214 9.984204e-01 2
## TICRR.2 2.728429e-05 -1.6652175 0.002 0.029 9.986595e-01 2
## LINC01215 2.749050e-05 0.7040675 0.030 0.077 1.000000e+00 2
## AP4M1.1 2.751899e-05 0.8561188 0.017 0.055 1.000000e+00 2
## GALNT7.1 2.791917e-05 1.2405361 0.075 0.145 1.000000e+00 2
## RNASEL 2.826848e-05 -0.2779709 0.015 0.053 1.000000e+00 2
## ZNF317 2.827206e-05 0.4114475 0.032 0.079 1.000000e+00 2
## FICD.1 2.827732e-05 0.7840972 0.011 0.045 1.000000e+00 2
## NEMP1.2 2.833828e-05 0.3490502 0.027 0.072 1.000000e+00 2
## METTL2A 2.846261e-05 0.2070023 0.015 0.053 1.000000e+00 2
## ATPSCKMT.1 2.861720e-05 0.1569344 0.015 0.053 1.000000e+00 2
## RUFY3.1 2.868330e-05 1.0523571 0.051 0.109 1.000000e+00 2
## MZF1-AS1 2.878573e-05 0.7165864 0.026 0.069 1.000000e+00 2
## TRAV13-2 2.896019e-05 -1.2941612 0.017 0.055 1.000000e+00 2
## ZNF764.1 2.910800e-05 0.5094421 0.014 0.050 1.000000e+00 2
## NHLRC3 2.911950e-05 0.7467908 0.029 0.074 1.000000e+00 2
## XPO1.1 2.919302e-05 0.8579155 0.200 0.343 1.000000e+00 2
## SPAST 2.929621e-05 0.6469089 0.054 0.113 1.000000e+00 2
## PEX3.1 2.932463e-05 0.5879540 0.042 0.095 1.000000e+00 2
## DYRK1A.1 2.959729e-05 1.9627027 0.281 0.273 1.000000e+00 2
## SUGP1 2.961124e-05 1.0506087 0.053 0.111 1.000000e+00 2
## SEPTIN11.1 2.967129e-05 0.9319410 0.078 0.149 1.000000e+00 2
## TMEM184C 2.969481e-05 0.9419691 0.044 0.097 1.000000e+00 2
## SPHK2 2.985749e-05 1.0373878 0.026 0.070 1.000000e+00 2
## MAP3K12 3.004328e-05 0.8475050 0.038 0.088 1.000000e+00 2
## WDR75.1 3.017260e-05 0.9288234 0.083 0.156 1.000000e+00 2
## DHRS7B.2 3.034334e-05 0.9013033 0.047 0.102 1.000000e+00 2
## PIGO 3.037526e-05 0.4625334 0.018 0.058 1.000000e+00 2
## ANKRD26.1 3.049778e-05 1.1367534 0.078 0.149 1.000000e+00 2
## RBM15.1 3.051621e-05 0.8401049 0.059 0.119 1.000000e+00 2
## TRGV3.2 3.052486e-05 -0.1000672 0.015 0.053 1.000000e+00 2
## MAP9 3.054224e-05 -0.1272599 0.015 0.053 1.000000e+00 2
## TFEB 3.062857e-05 0.1043169 0.018 0.058 1.000000e+00 2
## PLCD1 3.066082e-05 0.3248573 0.032 0.079 1.000000e+00 2
## AP4B1 3.068895e-05 1.2106421 0.047 0.101 1.000000e+00 2
## TRAV9-2 3.095465e-05 -0.5643917 0.014 0.050 1.000000e+00 2
## TRAV12-3 3.097291e-05 -1.6721729 0.008 0.040 1.000000e+00 2
## TIAM1.2 3.099444e-05 1.3074340 0.102 0.185 1.000000e+00 2
## LEF1-AS1.1 3.109445e-05 -0.8069276 0.008 0.040 1.000000e+00 2
## DTX2 3.116367e-05 1.0774212 0.053 0.111 1.000000e+00 2
## ZNF124 3.126895e-05 0.7639704 0.024 0.067 1.000000e+00 2
## NIFK-AS1 3.128485e-05 0.6982692 0.030 0.076 1.000000e+00 2
## ESF1.1 3.144379e-05 1.2226591 0.080 0.151 1.000000e+00 2
## SPPL2B.1 3.171722e-05 0.9877555 0.041 0.092 1.000000e+00 2
## PIAS3 3.192829e-05 0.3495761 0.020 0.060 1.000000e+00 2
## LINC01137 3.210620e-05 -1.0491415 0.005 0.034 1.000000e+00 2
## TRMT5 3.212421e-05 -1.2328536 0.003 0.031 1.000000e+00 2
## RFC3.2 3.213342e-05 0.9103487 0.035 0.083 1.000000e+00 2
## PHF2 3.215780e-05 0.7425272 0.044 0.097 1.000000e+00 2
## ZNF100.1 3.218472e-05 0.6921769 0.036 0.085 1.000000e+00 2
## GPRIN3.1 3.228887e-05 1.9231460 0.335 0.360 1.000000e+00 2
## PLEKHA7 3.245661e-05 -0.7724188 0.003 0.031 1.000000e+00 2
## PURB 3.246371e-05 0.9148056 0.059 0.119 1.000000e+00 2
## SSBP3.1 3.273240e-05 1.0196036 0.123 0.217 1.000000e+00 2
## KNOP1 3.293428e-05 1.0905225 0.042 0.094 1.000000e+00 2
## ARID4A 3.298939e-05 1.0915047 0.158 0.273 1.000000e+00 2
## HEATR3 3.298969e-05 0.4567267 0.038 0.087 1.000000e+00 2
## CNTROB.1 3.308759e-05 -0.4803800 0.008 0.040 1.000000e+00 2
## IPO9.2 3.328212e-05 1.0750210 0.084 0.157 1.000000e+00 2
## GINS1.2 3.330246e-05 0.9621431 0.014 0.050 1.000000e+00 2
## PLD6 3.333260e-05 -0.6748577 0.006 0.037 1.000000e+00 2
## RABL2B 3.334386e-05 -0.1758031 0.006 0.037 1.000000e+00 2
## SMIM13.1 3.343190e-05 0.7092324 0.023 0.064 1.000000e+00 2
## ZCCHC3 3.346297e-05 0.3401116 0.021 0.062 1.000000e+00 2
## FGFR1.1 3.348814e-05 0.7526873 0.044 0.096 1.000000e+00 2
## CEP44 3.353958e-05 0.7771117 0.035 0.083 1.000000e+00 2
## ZNF445 3.364442e-05 1.0303157 0.039 0.090 1.000000e+00 2
## HPS5 3.384506e-05 1.0802321 0.051 0.108 1.000000e+00 2
## ABHD4 3.388303e-05 -0.4451854 0.011 0.045 1.000000e+00 2
## POM121.1 3.404030e-05 1.1517080 0.108 0.195 1.000000e+00 2
## LMAN2L 3.434312e-05 0.3516238 0.011 0.045 1.000000e+00 2
## RPP40 3.454418e-05 -0.4736421 0.005 0.034 1.000000e+00 2
## BSDC1 3.461461e-05 0.9987789 0.072 0.139 1.000000e+00 2
## GNL3L.1 3.486781e-05 0.9874352 0.053 0.110 1.000000e+00 2
## THAP5 3.493649e-05 0.7538419 0.030 0.076 1.000000e+00 2
## DTX3 3.516066e-05 0.1595068 0.015 0.052 1.000000e+00 2
## ATG5 3.521684e-05 1.1124782 0.173 0.298 1.000000e+00 2
## PPP4R1 3.532195e-05 0.8033158 0.027 0.071 1.000000e+00 2
## ZKSCAN1 3.547606e-05 0.9429054 0.143 0.248 1.000000e+00 2
## AC015871.1.1 3.552854e-05 0.9312249 0.035 0.083 1.000000e+00 2
## TRAV12-1 3.562728e-05 -1.5163213 0.008 0.039 1.000000e+00 2
## ZNF10 3.566665e-05 0.7126031 0.048 0.103 1.000000e+00 2
## MX2.2 3.569770e-05 0.4500009 0.035 0.082 1.000000e+00 2
## SENP5.1 3.608880e-05 0.9761948 0.098 0.177 1.000000e+00 2
## ACOX1 3.620250e-05 0.8063854 0.042 0.094 1.000000e+00 2
## ZNF276.1 3.635860e-05 1.3361387 0.099 0.181 1.000000e+00 2
## NAXD 3.673663e-05 0.7710884 0.035 0.083 1.000000e+00 2
## ZNF33A 3.701718e-05 1.0035876 0.098 0.178 1.000000e+00 2
## NET1.2 3.715583e-05 0.4545385 0.023 0.064 1.000000e+00 2
## KCTD6 3.717467e-05 0.4476446 0.012 0.047 1.000000e+00 2
## LINC02361 3.717725e-05 0.3296471 0.021 0.062 1.000000e+00 2
## C1orf109 3.728312e-05 0.2254987 0.017 0.055 1.000000e+00 2
## ASCC3 3.732445e-05 1.7216114 0.379 0.435 1.000000e+00 2
## RIPK3.1 3.734279e-05 0.6347130 0.032 0.078 1.000000e+00 2
## SON 3.753006e-05 0.8643648 0.335 0.599 1.000000e+00 2
## ZFAND5 3.760801e-05 1.0916706 0.173 0.297 1.000000e+00 2
## NAPEPLD.1 3.763596e-05 0.3197194 0.015 0.052 1.000000e+00 2
## WIPI1.1 3.769539e-05 1.0605297 0.041 0.092 1.000000e+00 2
## SMAGP 3.773390e-05 0.1918265 0.009 0.042 1.000000e+00 2
## BPGM.1 3.788491e-05 0.8375535 0.023 0.064 1.000000e+00 2
## TOP3A.2 3.790372e-05 0.6153640 0.029 0.073 1.000000e+00 2
## C10orf88 3.810822e-05 0.6788073 0.023 0.064 1.000000e+00 2
## KANSL2.1 3.842523e-05 1.1303210 0.069 0.135 1.000000e+00 2
## KLF16 3.893877e-05 0.3290448 0.015 0.052 1.000000e+00 2
## COMMD10.1 3.899655e-05 1.1249369 0.177 0.305 1.000000e+00 2
## STXBP3 3.912450e-05 1.2251120 0.084 0.157 1.000000e+00 2
## MAP1LC3B2 3.933460e-05 -2.1414395 0.003 0.031 1.000000e+00 2
## MMS19.1 3.935381e-05 0.7919172 0.080 0.150 1.000000e+00 2
## GTF3C4 3.972683e-05 0.6063699 0.033 0.080 1.000000e+00 2
## AL157938.2 3.979942e-05 -1.0143352 0.003 0.031 1.000000e+00 2
## NINJ2.1 3.986174e-05 0.1101217 0.014 0.049 1.000000e+00 2
## GSEC 3.996040e-05 -0.8418266 0.003 0.031 1.000000e+00 2
## SLC12A9 4.021474e-05 0.4192931 0.038 0.087 1.000000e+00 2
## NDRG3.1 4.021505e-05 1.1814864 0.060 0.121 1.000000e+00 2
## ZNF407.1 4.055360e-05 2.2373302 0.220 0.191 1.000000e+00 2
## PIGW.1 4.064397e-05 -0.7542968 0.006 0.036 1.000000e+00 2
## LYZ.2 4.069323e-05 -4.2108828 0.002 0.028 1.000000e+00 2
## CEP85L.1 4.094962e-05 1.7632623 0.359 0.406 1.000000e+00 2
## SUCO.2 4.104967e-05 1.1186007 0.132 0.231 1.000000e+00 2
## PRMT5-AS1 4.135373e-05 3.4875023 0.038 0.016 1.000000e+00 2
## HIST1H1E 4.169906e-05 0.6695022 0.322 0.546 1.000000e+00 2
## PTPRN2.1 4.231786e-05 0.5899982 0.020 0.059 1.000000e+00 2
## MECP2.1 4.264122e-05 1.0817017 0.131 0.228 1.000000e+00 2
## MPP7-DT 4.267337e-05 4.3891565 0.014 0.003 1.000000e+00 2
## ATG9B.1 4.278124e-05 -0.5385075 0.003 0.031 1.000000e+00 2
## LINC00539 4.286230e-05 0.1875714 0.021 0.061 1.000000e+00 2
## FAR1 4.288471e-05 1.0787835 0.105 0.188 1.000000e+00 2
## NUP50-DT 4.299493e-05 0.6634752 0.020 0.059 1.000000e+00 2
## ZBTB6 4.312165e-05 0.4521898 0.018 0.056 1.000000e+00 2
## PHF7.1 4.320005e-05 0.9083575 0.020 0.059 1.000000e+00 2
## AL133268.4 4.353337e-05 0.7548565 0.026 0.068 1.000000e+00 2
## TRAV12-2.2 4.354298e-05 -0.9091423 0.020 0.058 1.000000e+00 2
## PRKD3 4.356487e-05 0.2404869 0.023 0.063 1.000000e+00 2
## SLAMF7.1 4.406888e-05 0.6170354 0.069 0.132 1.000000e+00 2
## ZNHIT6.1 4.414922e-05 1.2067331 0.075 0.143 1.000000e+00 2
## IPO13.1 4.445930e-05 0.2776053 0.017 0.054 1.000000e+00 2
## Z94721.1.2 4.477189e-05 0.2543805 0.018 0.056 1.000000e+00 2
## A1BG-AS1 4.483198e-05 -0.4459383 0.014 0.049 1.000000e+00 2
## HSDL1 4.487733e-05 0.3589759 0.014 0.049 1.000000e+00 2
## FOXN2 4.498894e-05 1.0883632 0.168 0.288 1.000000e+00 2
## ZSCAN16 4.501490e-05 0.1893924 0.017 0.054 1.000000e+00 2
## PEX14 4.515438e-05 1.1236266 0.089 0.163 1.000000e+00 2
## CTSF 4.546292e-05 -1.3016127 0.005 0.033 1.000000e+00 2
## SEC24D 4.552242e-05 1.1654286 0.110 0.195 1.000000e+00 2
## APP.2 4.585277e-05 0.8610281 0.051 0.107 1.000000e+00 2
## SYNJ2BP 4.617448e-05 0.8330117 0.024 0.066 1.000000e+00 2
## KLHL42.2 4.626273e-05 0.5955657 0.035 0.082 1.000000e+00 2
## MSTO1.1 4.654427e-05 0.8804107 0.036 0.084 1.000000e+00 2
## LYL1.1 4.665036e-05 -0.2409294 0.006 0.036 1.000000e+00 2
## CHTF18.2 4.667631e-05 0.5368460 0.017 0.054 1.000000e+00 2
## BICD2 4.668222e-05 1.1714742 0.057 0.116 1.000000e+00 2
## GABPB1.1 4.671798e-05 1.0141337 0.065 0.127 1.000000e+00 2
## MATR3.1 4.694902e-05 0.7292072 0.054 0.111 1.000000e+00 2
## NOP14-AS1 4.706736e-05 0.8897431 0.035 0.082 1.000000e+00 2
## ZC3H18.1 4.724858e-05 1.1837740 0.126 0.221 1.000000e+00 2
## CARD11.1 4.737953e-05 1.9750471 0.329 0.353 1.000000e+00 2
## AC025171.1 4.763449e-05 0.9974824 0.053 0.109 1.000000e+00 2
## LTO1 4.799473e-05 0.5234257 0.023 0.063 1.000000e+00 2
## GATAD1 4.819626e-05 0.4614706 0.036 0.084 1.000000e+00 2
## ALMS1.1 4.835785e-05 1.1548875 0.071 0.136 1.000000e+00 2
## DHCR24.2 4.861645e-05 0.7338816 0.021 0.061 1.000000e+00 2
## TMEM237.2 4.867526e-05 -1.1793577 0.003 0.030 1.000000e+00 2
## NFKBIE.1 4.878440e-05 0.6480773 0.026 0.068 1.000000e+00 2
## STK40.1 4.955382e-05 0.9746126 0.065 0.127 1.000000e+00 2
## SELENON.2 5.012265e-05 -4.2305303 0.000 0.024 1.000000e+00 2
## FBXO25 5.012532e-05 0.3616528 0.018 0.056 1.000000e+00 2
## FAM160B2 5.026192e-05 0.2101989 0.024 0.065 1.000000e+00 2
## CD248.1 5.026234e-05 -1.1484820 0.009 0.041 1.000000e+00 2
## RMND5A.1 5.028649e-05 1.2313493 0.098 0.176 1.000000e+00 2
## PLEKHM2.1 5.035189e-05 1.1133665 0.059 0.118 1.000000e+00 2
## SECISBP2 5.047650e-05 1.0431831 0.138 0.240 1.000000e+00 2
## CSGALNACT2 5.049259e-05 1.1370698 0.159 0.272 1.000000e+00 2
## LMNA.1 5.071896e-05 1.1222714 0.054 0.111 1.000000e+00 2
## TRBV19 5.073756e-05 -1.6932929 0.023 0.061 1.000000e+00 2
## TWSG1 5.076254e-05 0.6000444 0.023 0.063 1.000000e+00 2
## ARHGEF39.2 5.081247e-05 -2.7596124 0.002 0.027 1.000000e+00 2
## ZRSR2 5.099321e-05 1.1431168 0.057 0.115 1.000000e+00 2
## ZBTB11 5.130099e-05 1.0010969 0.126 0.220 1.000000e+00 2
## PHF12.1 5.133346e-05 0.9915974 0.081 0.151 1.000000e+00 2
## XRCC3.2 5.151578e-05 1.1880499 0.027 0.070 1.000000e+00 2
## ALOX12-AS1 5.188272e-05 0.8285868 0.051 0.106 1.000000e+00 2
## KBTBD3 5.202697e-05 0.6190091 0.027 0.070 1.000000e+00 2
## AL390066.1 5.227692e-05 3.5403054 0.017 0.004 1.000000e+00 2
## EPSTI1.2 5.241530e-05 1.1892410 0.078 0.147 1.000000e+00 2
## STRN4 5.254325e-05 0.6269656 0.042 0.092 1.000000e+00 2
## KIFAP3 5.281694e-05 1.1573767 0.113 0.199 1.000000e+00 2
## INTS2 5.313577e-05 0.6788422 0.029 0.072 1.000000e+00 2
## CRNDE.2 5.330185e-05 0.1568675 0.008 0.038 1.000000e+00 2
## STAT6 5.339854e-05 1.2464257 0.114 0.202 1.000000e+00 2
## ODF2L.1 5.341161e-05 1.1198262 0.161 0.276 1.000000e+00 2
## PDPK1 5.362907e-05 1.1105281 0.056 0.113 1.000000e+00 2
## ZFP62 5.413940e-05 0.1629080 0.015 0.051 1.000000e+00 2
## LZTS2 5.441105e-05 0.5193032 0.024 0.065 1.000000e+00 2
## CENPJ.2 5.447345e-05 1.1269908 0.030 0.074 1.000000e+00 2
## NEK1.1 5.466576e-05 1.3221335 0.137 0.237 1.000000e+00 2
## APOBEC3D 5.518544e-05 0.4632022 0.026 0.067 1.000000e+00 2
## PNPLA8 5.525365e-05 1.0073560 0.092 0.167 1.000000e+00 2
## VPS35L 5.559344e-05 0.9813508 0.087 0.159 1.000000e+00 2
## UTP20.1 5.573414e-05 1.2870185 0.044 0.095 1.000000e+00 2
## NBPF19.1 5.669178e-05 1.0990410 0.048 0.101 1.000000e+00 2
## METTL4 5.691388e-05 1.0472346 0.036 0.083 1.000000e+00 2
## AKAP8L.1 5.702546e-05 1.2842592 0.123 0.215 1.000000e+00 2
## PUDP 5.733042e-05 0.6566659 0.039 0.088 1.000000e+00 2
## LRRC37A3 5.742674e-05 0.1851889 0.015 0.051 1.000000e+00 2
## WDR92 5.777838e-05 0.3372900 0.011 0.043 1.000000e+00 2
## ITGA4.2 5.778562e-05 1.0968911 0.532 0.735 1.000000e+00 2
## ZNF430.1 5.798648e-05 1.2570494 0.086 0.157 1.000000e+00 2
## CENPP.2 5.833338e-05 0.9145137 0.089 0.160 1.000000e+00 2
## ACTR8 5.833364e-05 1.0175040 0.039 0.088 1.000000e+00 2
## COQ3 5.838708e-05 0.4939132 0.012 0.046 1.000000e+00 2
## FHL1.1 5.847317e-05 0.3411055 0.012 0.046 1.000000e+00 2
## CMSS1.2 5.848736e-05 1.0054826 0.065 0.126 1.000000e+00 2
## OSBPL11 5.898326e-05 0.5242580 0.033 0.079 1.000000e+00 2
## NMT2.1 5.904278e-05 1.2352070 0.098 0.176 1.000000e+00 2
## TRIM26 5.904436e-05 1.0751522 0.105 0.187 1.000000e+00 2
## TTC21B 5.906496e-05 0.6719115 0.039 0.088 1.000000e+00 2
## RNF146 5.945008e-05 0.5981196 0.041 0.090 1.000000e+00 2
## USP4 5.964202e-05 1.1791075 0.140 0.240 1.000000e+00 2
## MAPKAPK2.1 6.008557e-05 1.1063474 0.086 0.158 1.000000e+00 2
## PER1.1 6.032599e-05 1.1743484 0.119 0.208 1.000000e+00 2
## SYNJ2 6.110115e-05 1.2344607 0.072 0.137 1.000000e+00 2
## ZNF557.1 6.112497e-05 0.7393597 0.024 0.065 1.000000e+00 2
## ANGEL1 6.136059e-05 -0.1454455 0.011 0.043 1.000000e+00 2
## PGBD2 6.170784e-05 -0.1089489 0.012 0.046 1.000000e+00 2
## THAP8 6.178938e-05 0.7683180 0.011 0.043 1.000000e+00 2
## TMX3 6.243998e-05 1.1291060 0.114 0.201 1.000000e+00 2
## APOL6.1 6.316396e-05 1.1122873 0.179 0.303 1.000000e+00 2
## RNF40 6.332901e-05 0.9842462 0.059 0.117 1.000000e+00 2
## METTL2B 6.336195e-05 0.8219783 0.024 0.065 1.000000e+00 2
## AC067945.4 6.337033e-05 0.1843290 0.018 0.055 1.000000e+00 2
## TRIP4 6.340589e-05 0.8082868 0.056 0.112 1.000000e+00 2
## NFS1.1 6.347422e-05 1.0369735 0.030 0.074 1.000000e+00 2
## AP3S2 6.354695e-05 1.1292022 0.053 0.108 1.000000e+00 2
## RCBTB1.1 6.380699e-05 0.8084099 0.041 0.090 1.000000e+00 2
## PICK1 6.394458e-05 0.4565812 0.009 0.040 1.000000e+00 2
## RARG 6.398757e-05 0.9380087 0.039 0.087 1.000000e+00 2
## MFAP3 6.447813e-05 0.3135976 0.020 0.058 1.000000e+00 2
## DNAJA4.1 6.453605e-05 -1.3539353 0.006 0.035 1.000000e+00 2
## FAIM 6.468981e-05 -0.9809019 0.003 0.030 1.000000e+00 2
## CDK6.1 6.477524e-05 1.5512512 0.441 0.560 1.000000e+00 2
## BPHL.1 6.536237e-05 -0.3597438 0.006 0.035 1.000000e+00 2
## FURIN.1 6.537234e-05 1.0752984 0.093 0.168 1.000000e+00 2
## REV1.2 6.560680e-05 1.0762227 0.084 0.154 1.000000e+00 2
## TPD52.1 6.560949e-05 1.2515454 0.155 0.265 1.000000e+00 2
## SPAG1 6.576555e-05 0.3890133 0.021 0.060 1.000000e+00 2
## ZNF518B.1 6.600295e-05 0.3864619 0.026 0.067 1.000000e+00 2
## MPHOSPH9.1 6.618692e-05 1.1506080 0.114 0.199 1.000000e+00 2
## ASTE1 6.640359e-05 1.0131664 0.041 0.090 1.000000e+00 2
## EDEM3 6.642545e-05 1.2651445 0.135 0.233 1.000000e+00 2
## PARP12 6.684858e-05 1.1238965 0.068 0.130 1.000000e+00 2
## AC006369.1.2 6.703011e-05 0.2966025 0.036 0.083 1.000000e+00 2
## CCDC150.2 6.725695e-05 -1.9385349 0.002 0.026 1.000000e+00 2
## TRIM56 6.753917e-05 1.3954076 0.093 0.168 1.000000e+00 2
## CYTH1 6.776702e-05 1.4963536 0.426 0.537 1.000000e+00 2
## SLC25A42 6.800950e-05 1.1685393 0.042 0.092 1.000000e+00 2
## AL360178.1 6.809522e-05 -1.2568740 0.005 0.032 1.000000e+00 2
## TRMT12 6.825854e-05 0.7832520 0.017 0.053 1.000000e+00 2
## ALDH1B1 6.833305e-05 -0.7549801 0.005 0.032 1.000000e+00 2
## CD200R1.1 6.833462e-05 1.0624602 0.054 0.110 1.000000e+00 2
## IRF5 6.840454e-05 -0.1024556 0.005 0.032 1.000000e+00 2
## NFATC3 6.848129e-05 1.6725022 0.395 0.476 1.000000e+00 2
## HIST1H2AH.2 6.851291e-05 0.5053238 0.032 0.076 1.000000e+00 2
## SLC35E3 6.858509e-05 1.1271977 0.026 0.067 1.000000e+00 2
## TRAV8-3 6.873398e-05 0.1285855 0.020 0.057 1.000000e+00 2
## HERC5 6.889226e-05 0.5470921 0.035 0.080 1.000000e+00 2
## JADE1.1 6.934469e-05 1.0013011 0.086 0.156 1.000000e+00 2
## GSTM3.1 6.938744e-05 0.6549998 0.011 0.043 1.000000e+00 2
## IP6K1 6.943101e-05 1.2817931 0.089 0.161 1.000000e+00 2
## PPP3CB-AS1 7.050252e-05 0.6261538 0.024 0.064 1.000000e+00 2
## STAG3.2 7.085483e-05 0.6522873 0.014 0.048 1.000000e+00 2
## NEK4 7.167620e-05 0.5960671 0.021 0.060 1.000000e+00 2
## FLCN 7.182518e-05 0.9242582 0.036 0.083 1.000000e+00 2
## IDH1 7.209866e-05 1.1322575 0.021 0.060 1.000000e+00 2
## VANGL1.1 7.216371e-05 0.1227898 0.011 0.043 1.000000e+00 2
## HLA-G 7.226575e-05 -0.6484209 0.002 0.026 1.000000e+00 2
## AQR.1 7.246201e-05 1.0653365 0.102 0.182 1.000000e+00 2
## PPFIA1 7.317044e-05 1.0415840 0.096 0.172 1.000000e+00 2
## ZNF512 7.326681e-05 0.9481350 0.063 0.122 1.000000e+00 2
## THUMPD2.1 7.350023e-05 1.0021310 0.060 0.118 1.000000e+00 2
## ZNF726.1 7.367861e-05 -0.7905021 0.003 0.029 1.000000e+00 2
## TRAV39 7.383964e-05 -0.9426450 0.006 0.035 1.000000e+00 2
## PET117 7.409920e-05 -0.7844566 0.006 0.035 1.000000e+00 2
## ZYG11B.1 7.427978e-05 0.9607157 0.033 0.078 1.000000e+00 2
## ZNF140 7.435007e-05 0.6519566 0.026 0.066 1.000000e+00 2
## SMIM8 7.473176e-05 0.5796037 0.021 0.059 1.000000e+00 2
## ZNF684.1 7.489558e-05 0.4623926 0.006 0.035 1.000000e+00 2
## METTL16.1 7.514269e-05 1.1436149 0.069 0.132 1.000000e+00 2
## CPT2 7.555029e-05 0.1214114 0.014 0.047 1.000000e+00 2
## DNM1L.1 7.602267e-05 1.1831374 0.108 0.190 1.000000e+00 2
## RRAS2.2 7.611437e-05 1.0521269 0.113 0.197 1.000000e+00 2
## FAM161A.2 7.618890e-05 -4.4801645 0.000 0.023 1.000000e+00 2
## CC2D1A.1 7.675756e-05 1.0636592 0.047 0.098 1.000000e+00 2
## SCAP 7.696356e-05 0.9829065 0.071 0.134 1.000000e+00 2
## BRD9.1 7.709147e-05 1.4400158 0.057 0.114 1.000000e+00 2
## USP15 7.812932e-05 1.4258917 0.450 0.592 1.000000e+00 2
## GNL3.2 7.916403e-05 1.2887858 0.093 0.168 1.000000e+00 2
## ORC2.1 7.926292e-05 1.1073179 0.087 0.158 1.000000e+00 2
## AMACR 7.928568e-05 0.3997838 0.011 0.042 1.000000e+00 2
## FKBPL.1 7.942387e-05 -1.4078266 0.003 0.029 1.000000e+00 2
## PRMT6 7.974937e-05 0.1248614 0.020 0.057 1.000000e+00 2
## LUC7L 7.988531e-05 1.2451451 0.110 0.193 1.000000e+00 2
## MIOS 7.995762e-05 0.8777229 0.039 0.087 1.000000e+00 2
## KYAT1 8.006842e-05 0.2519202 0.018 0.055 1.000000e+00 2
## SGCB 8.010177e-05 -0.1180442 0.006 0.034 1.000000e+00 2
## AL356356.1 8.141011e-05 4.2997546 0.014 0.003 1.000000e+00 2
## ABHD5 8.178687e-05 1.2257025 0.045 0.095 1.000000e+00 2
## ZNF260 8.182589e-05 -0.7333727 0.008 0.037 1.000000e+00 2
## STAT2.1 8.233195e-05 1.2383765 0.087 0.158 1.000000e+00 2
## BRMS1L 8.233490e-05 -0.1428835 0.009 0.040 1.000000e+00 2
## TTPAL 8.241713e-05 0.6788575 0.023 0.062 1.000000e+00 2
## ZNF232.2 8.248657e-05 -0.5363025 0.005 0.032 1.000000e+00 2
## TRAV13-1.1 8.252191e-05 -1.0446614 0.033 0.075 1.000000e+00 2
## TRBV27.1 8.258151e-05 -1.3898185 0.017 0.051 1.000000e+00 2
## ZNF599 8.298131e-05 -2.2267463 0.002 0.026 1.000000e+00 2
## GFM2.1 8.321570e-05 0.4918143 0.035 0.080 1.000000e+00 2
## ZW10.1 8.380080e-05 1.0512540 0.029 0.071 1.000000e+00 2
## BIRC3.2 8.420039e-05 1.0879388 0.152 0.256 1.000000e+00 2
## CD6 8.493732e-05 0.8794148 0.295 0.508 1.000000e+00 2
## ICK 8.494889e-05 0.1255475 0.017 0.052 1.000000e+00 2
## SLC30A7 8.501096e-05 1.1484337 0.126 0.218 1.000000e+00 2
## ZNF691 8.513838e-05 0.1012999 0.008 0.037 1.000000e+00 2
## ZNF395 8.527311e-05 -0.7870825 0.003 0.029 1.000000e+00 2
## UBXN8.1 8.537024e-05 0.4796762 0.014 0.047 1.000000e+00 2
## SDCCAG8 8.561500e-05 1.1258310 0.105 0.185 1.000000e+00 2
## MSL1 8.566175e-05 0.8707056 0.078 0.144 1.000000e+00 2
## SLC2A11 8.686137e-05 0.4341711 0.020 0.057 1.000000e+00 2
## TUBB4A.1 8.760492e-05 -4.7670644 0.000 0.023 1.000000e+00 2
## LOXL1-AS1.1 8.779217e-05 -0.9111559 0.006 0.034 1.000000e+00 2
## MACROD1.2 8.783006e-05 1.1409106 0.024 0.063 1.000000e+00 2
## SIDT2 8.805620e-05 -0.7354991 0.011 0.042 1.000000e+00 2
## ACOX3.1 8.822379e-05 0.7428758 0.021 0.059 1.000000e+00 2
## RLIM 8.833405e-05 1.1935417 0.102 0.180 1.000000e+00 2
## WAC 8.872885e-05 0.9531515 0.218 0.368 1.000000e+00 2
## TRAV41 8.878428e-05 -2.5541760 0.002 0.026 1.000000e+00 2
## CORO2A 8.891366e-05 0.8362677 0.035 0.080 1.000000e+00 2
## SPINT1.1 8.892121e-05 -2.1651301 0.002 0.026 1.000000e+00 2
## GAA 8.893097e-05 0.5104187 0.014 0.047 1.000000e+00 2
## CACUL1 8.939196e-05 1.0298068 0.068 0.129 1.000000e+00 2
## NAA40.1 8.987896e-05 0.5913124 0.033 0.077 1.000000e+00 2
## POLR3F 9.036133e-05 0.6553895 0.011 0.042 1.000000e+00 2
## TXLNA.1 9.038880e-05 1.1965932 0.075 0.140 1.000000e+00 2
## ZDHHC14.2 9.039022e-05 2.6003501 0.158 0.122 1.000000e+00 2
## PANX1 9.056560e-05 0.6393547 0.018 0.054 1.000000e+00 2
## GPR160.1 9.075131e-05 -0.3294749 0.008 0.037 1.000000e+00 2
## ABHD3 9.120074e-05 1.1644104 0.140 0.238 1.000000e+00 2
## TRAPPC9.1 9.124710e-05 2.0889783 0.272 0.265 1.000000e+00 2
## ACBD4 9.155056e-05 0.5251025 0.012 0.044 1.000000e+00 2
## ZNF107.2 9.160455e-05 1.0309993 0.077 0.142 1.000000e+00 2
## PPP2CB 9.228270e-05 1.0485592 0.032 0.075 1.000000e+00 2
## SIRT5.1 9.229111e-05 0.5858581 0.021 0.059 1.000000e+00 2
## SLC11A2.1 9.243536e-05 0.6782271 0.036 0.082 1.000000e+00 2
## ASB2.1 9.262051e-05 1.0976281 0.161 0.270 1.000000e+00 2
## AC005944.1 9.394011e-05 -4.6472551 0.000 0.022 1.000000e+00 2
## ERCC6L.2 9.397387e-05 -3.4591712 0.002 0.026 1.000000e+00 2
## PLCB2.1 9.421113e-05 1.2541758 0.110 0.192 1.000000e+00 2
## AL392172.1 9.450125e-05 0.1313870 0.009 0.039 1.000000e+00 2
## PGAP3 9.457922e-05 0.7353579 0.017 0.052 1.000000e+00 2
## TRIM34 9.473554e-05 -0.2052838 0.009 0.039 1.000000e+00 2
## FBXL8.1 9.475172e-05 1.1820217 0.063 0.122 1.000000e+00 2
## ROCK2.1 9.495852e-05 1.1325904 0.092 0.164 1.000000e+00 2
## RGP1 9.524186e-05 0.6512988 0.032 0.075 1.000000e+00 2
## SCMH1.2 9.526282e-05 1.0699438 0.080 0.145 1.000000e+00 2
## SPATA33.1 9.536488e-05 0.5278630 0.026 0.066 1.000000e+00 2
## AKNA.1 9.612504e-05 1.6664936 0.388 0.461 1.000000e+00 2
## ZNF200 9.635836e-05 0.3020241 0.015 0.049 1.000000e+00 2
## ZNF211 9.648849e-05 0.3857681 0.018 0.054 1.000000e+00 2
## USP19 9.694558e-05 0.5885448 0.030 0.072 1.000000e+00 2
## MAF.2 9.697672e-05 1.1291463 0.072 0.135 1.000000e+00 2
## SLC27A4 9.733965e-05 0.9703902 0.023 0.061 1.000000e+00 2
## NRDC.1 9.743603e-05 1.1036554 0.236 0.403 1.000000e+00 2
## FBXW9 9.772194e-05 -0.1984943 0.003 0.028 1.000000e+00 2
## EFCAB2.1 9.775789e-05 1.2132605 0.068 0.128 1.000000e+00 2
## KLHDC10 9.779718e-05 1.0075194 0.068 0.128 1.000000e+00 2
## ESR2 9.787172e-05 0.7439610 0.042 0.091 1.000000e+00 2
## ELF4 9.875878e-05 0.8053650 0.051 0.104 1.000000e+00 2
## SOCS4.1 9.939145e-05 1.0491213 0.056 0.110 1.000000e+00 2
## LEMD2 9.981274e-05 1.1161950 0.062 0.119 1.000000e+00 2
## PAN2 1.001318e-04 0.8381586 0.020 0.056 1.000000e+00 2
## KMT2B 1.003157e-04 1.3246333 0.114 0.198 1.000000e+00 2
## RTN4IP1 1.006896e-04 0.2547039 0.009 0.039 1.000000e+00 2
## PCM1 1.008866e-04 1.0954140 0.254 0.435 1.000000e+00 2
## ZNF701 1.010503e-04 0.5802418 0.026 0.065 1.000000e+00 2
## TBX21.2 1.015456e-04 0.7503180 0.068 0.127 1.000000e+00 2
## FAM153CP 1.022258e-04 -2.2994936 0.002 0.025 1.000000e+00 2
## COA7.1 1.026580e-04 0.3102385 0.011 0.042 1.000000e+00 2
## CCNG2.1 1.028920e-04 0.5865323 0.047 0.096 1.000000e+00 2
## ZNF275 1.029895e-04 0.3013803 0.014 0.047 1.000000e+00 2
## STRIP1 1.033343e-04 0.9628925 0.051 0.103 1.000000e+00 2
## SLC35F2.1 1.034325e-04 0.6969646 0.006 0.034 1.000000e+00 2
## C9orf72.1 1.042043e-04 1.2098531 0.051 0.104 1.000000e+00 2
## PCTP 1.045352e-04 -1.7346958 0.003 0.028 1.000000e+00 2
## FAF1.1 1.048183e-04 1.6601206 0.359 0.407 1.000000e+00 2
## SPIN1.1 1.052638e-04 0.9987569 0.053 0.106 1.000000e+00 2
## TRAV22 1.053328e-04 -0.5889262 0.011 0.041 1.000000e+00 2
## COPG2.2 1.059655e-04 0.5683276 0.020 0.056 1.000000e+00 2
## ARHGAP33.2 1.059952e-04 -0.1887265 0.009 0.039 1.000000e+00 2
## ZNF286A 1.061062e-04 0.1910158 0.015 0.049 1.000000e+00 2
## ZBTB22 1.064875e-04 -0.4224649 0.009 0.039 1.000000e+00 2
## THAP2 1.066508e-04 -0.1949632 0.012 0.044 1.000000e+00 2
## ERLIN2.1 1.072752e-04 0.7018857 0.027 0.067 1.000000e+00 2
## FOXP1.1 1.075847e-04 1.5752792 0.423 0.531 1.000000e+00 2
## CLSTN3.1 1.076743e-04 1.1533663 0.072 0.135 1.000000e+00 2
## TRAV25 1.080203e-04 -5.9608215 0.000 0.022 1.000000e+00 2
## RAB33B 1.081129e-04 0.4497965 0.023 0.061 1.000000e+00 2
## SLA 1.083671e-04 1.1843919 0.137 0.233 1.000000e+00 2
## DLG3 1.084259e-04 0.8502215 0.021 0.058 1.000000e+00 2
## HDAC7.1 1.084287e-04 1.4184149 0.092 0.164 1.000000e+00 2
## TEFM 1.084437e-04 0.7848718 0.030 0.072 1.000000e+00 2
## GORAB 1.090712e-04 0.7189926 0.015 0.049 1.000000e+00 2
## POMC 1.096706e-04 -1.3857393 0.002 0.025 1.000000e+00 2
## ABCC10 1.103105e-04 1.0112721 0.035 0.079 1.000000e+00 2
## FBXO21 1.105075e-04 0.9651444 0.074 0.137 1.000000e+00 2
## INF2.1 1.118707e-04 1.2576677 0.056 0.110 1.000000e+00 2
## ANKRD49 1.119321e-04 1.0157170 0.026 0.065 1.000000e+00 2
## RRNAD1 1.125177e-04 0.9844894 0.030 0.072 1.000000e+00 2
## PEX1 1.128873e-04 0.8786618 0.050 0.101 1.000000e+00 2
## KMT5C 1.137545e-04 1.0024580 0.027 0.067 1.000000e+00 2
## CCDC106.1 1.139576e-04 -0.6845528 0.005 0.031 1.000000e+00 2
## ARSK.1 1.143406e-04 0.8361073 0.021 0.058 1.000000e+00 2
## ZNF181 1.145091e-04 -0.3289614 0.009 0.039 1.000000e+00 2
## TRAV36DV7 1.146711e-04 -1.3235540 0.005 0.031 1.000000e+00 2
## MFSD14B.1 1.148696e-04 1.0414759 0.075 0.139 1.000000e+00 2
## ZDHHC13.1 1.153996e-04 1.2741161 0.065 0.123 1.000000e+00 2
## RCOR3.1 1.163173e-04 1.0132572 0.107 0.186 1.000000e+00 2
## HPGD.1 1.167484e-04 0.2576790 0.020 0.056 1.000000e+00 2
## MOV10.2 1.169373e-04 0.7067100 0.029 0.070 1.000000e+00 2
## DNAJB2 1.174409e-04 1.2853407 0.041 0.088 1.000000e+00 2
## CRACR2B.1 1.176577e-04 0.4107324 0.011 0.041 1.000000e+00 2
## TOB2 1.177517e-04 1.1346668 0.081 0.148 1.000000e+00 2
## AK5.1 1.178120e-04 -0.6759181 0.005 0.031 1.000000e+00 2
## FASN.2 1.178394e-04 1.0219646 0.045 0.094 1.000000e+00 2
## URGCP.1 1.187732e-04 1.1499881 0.044 0.092 1.000000e+00 2
## DZIP3.1 1.189018e-04 0.9111703 0.072 0.134 1.000000e+00 2
## SPATA5L1 1.193343e-04 0.3917803 0.017 0.051 1.000000e+00 2
## CPNE7 1.199088e-04 -0.1363055 0.009 0.038 1.000000e+00 2
## SLC16A1.2 1.201998e-04 1.4956968 0.071 0.132 1.000000e+00 2
## GLYCTK 1.208985e-04 0.7623428 0.030 0.072 1.000000e+00 2
## PAOX.1 1.213721e-04 0.1249685 0.015 0.048 1.000000e+00 2
## CXorf56 1.219226e-04 -0.1824280 0.011 0.041 1.000000e+00 2
## ELF2 1.222049e-04 1.1590728 0.174 0.294 1.000000e+00 2
## ZAP70.2 1.226163e-04 0.9617694 0.337 0.593 1.000000e+00 2
## WDR24 1.228245e-04 0.2894774 0.009 0.038 1.000000e+00 2
## PAFAH1B1 1.229796e-04 0.9094471 0.266 0.456 1.000000e+00 2
## PPP3CB 1.229951e-04 1.2277161 0.126 0.216 1.000000e+00 2
## STK3.2 1.235540e-04 0.1863273 0.012 0.043 1.000000e+00 2
## TRIM32 1.236256e-04 0.8625293 0.015 0.048 1.000000e+00 2
## RASA3.1 1.237762e-04 1.0021592 0.259 0.437 1.000000e+00 2
## ZNF740 1.237961e-04 0.7958537 0.038 0.083 1.000000e+00 2
## NCAM1.2 1.247377e-04 3.3623321 0.027 0.010 1.000000e+00 2
## ATAD3B.1 1.249540e-04 1.0220203 0.027 0.067 1.000000e+00 2
## CENPBD1 1.252295e-04 -1.4779051 0.005 0.030 1.000000e+00 2
## CEP68 1.256395e-04 0.5633595 0.032 0.074 1.000000e+00 2
## ZNF724.1 1.257182e-04 0.4732837 0.012 0.043 1.000000e+00 2
## FAM86C1 1.261861e-04 0.2689347 0.005 0.030 1.000000e+00 2
## SIAH1.1 1.263327e-04 1.0721159 0.038 0.083 1.000000e+00 2
## SUGP2.1 1.265943e-04 1.2963312 0.089 0.158 1.000000e+00 2
## CTSO 1.268664e-04 -0.9841944 0.006 0.033 1.000000e+00 2
## YTHDC2.1 1.268699e-04 0.9875329 0.113 0.194 1.000000e+00 2
## AC012447.1.1 1.273418e-04 2.6044346 0.093 0.059 1.000000e+00 2
## SENP2 1.279197e-04 1.0118806 0.057 0.111 1.000000e+00 2
## AC147067.1.1 1.287029e-04 -0.9364106 0.003 0.028 1.000000e+00 2
## GSK3B.1 1.291967e-04 1.8870375 0.332 0.365 1.000000e+00 2
## DCP1B 1.293288e-04 0.6622312 0.033 0.076 1.000000e+00 2
## SC5D 1.298022e-04 1.1014569 0.035 0.078 1.000000e+00 2
## CAPN10 1.298551e-04 1.1933341 0.045 0.094 1.000000e+00 2
## CD247.1 1.302839e-04 0.7589907 0.361 0.632 1.000000e+00 2
## PLOD1.1 1.323663e-04 -0.5778777 0.008 0.036 1.000000e+00 2
## AC006449.6 1.326195e-04 -0.5544856 0.008 0.036 1.000000e+00 2
## KIAA1328.2 1.328979e-04 0.9681799 0.077 0.140 1.000000e+00 2
## DBF4B.2 1.331134e-04 -1.1186238 0.005 0.030 1.000000e+00 2
## SNHG11 1.331848e-04 0.1512070 0.011 0.041 1.000000e+00 2
## HELLPAR.2 1.332028e-04 -4.4064961 0.000 0.022 1.000000e+00 2
## FXR2 1.334120e-04 0.5273097 0.029 0.069 1.000000e+00 2
## VPS41 1.335563e-04 0.9728301 0.126 0.215 1.000000e+00 2
## TRIM44.2 1.336222e-04 1.1657433 0.087 0.156 1.000000e+00 2
## USP34 1.340070e-04 1.8175477 0.341 0.382 1.000000e+00 2
## PRR11.2 1.341485e-04 0.1895320 0.008 0.036 1.000000e+00 2
## GATB.1 1.341490e-04 0.8609438 0.027 0.067 1.000000e+00 2
## DYRK1B.1 1.350786e-04 0.7030898 0.026 0.064 1.000000e+00 2
## AC008035.1.1 1.351095e-04 -2.1062496 0.002 0.025 1.000000e+00 2
## FAM199X 1.352114e-04 0.7478795 0.038 0.083 1.000000e+00 2
## RARS2 1.355133e-04 1.1797286 0.104 0.180 1.000000e+00 2
## ANKRD13C 1.356144e-04 1.0323861 0.119 0.203 1.000000e+00 2
## UBL7-AS1.2 1.360138e-04 1.3688675 0.050 0.100 1.000000e+00 2
## ARIH2OS 1.367408e-04 0.6185532 0.020 0.055 1.000000e+00 2
## FANCC.2 1.369573e-04 1.3045887 0.033 0.076 1.000000e+00 2
## LRMP 1.377073e-04 0.9976599 0.066 0.124 1.000000e+00 2
## NUMA1 1.379268e-04 1.1942396 0.144 0.244 1.000000e+00 2
## CCDC159 1.387965e-04 0.7677767 0.018 0.053 1.000000e+00 2
## LTBP3.1 1.405207e-04 1.2422960 0.045 0.093 1.000000e+00 2
## NSRP1 1.405272e-04 1.2001861 0.132 0.224 1.000000e+00 2
## ZBTB7B.2 1.405497e-04 1.0847853 0.056 0.109 1.000000e+00 2
## SETD6 1.407555e-04 0.3339270 0.014 0.046 1.000000e+00 2
## TMEM175 1.412472e-04 1.0946408 0.053 0.104 1.000000e+00 2
## ZNF37A.1 1.412614e-04 1.2473698 0.096 0.169 1.000000e+00 2
## FRAT1 1.415583e-04 0.6404495 0.017 0.050 1.000000e+00 2
## FBXO38 1.417819e-04 1.1657398 0.075 0.138 1.000000e+00 2
## ANKRD27 1.432528e-04 1.1057197 0.075 0.138 1.000000e+00 2
## ZNF429 1.433318e-04 0.4625676 0.023 0.060 1.000000e+00 2
## LIN9.2 1.435752e-04 0.8379509 0.017 0.050 1.000000e+00 2
## RHOU.1 1.438393e-04 0.8099641 0.021 0.057 1.000000e+00 2
## MISP3 1.439164e-04 -1.0570231 0.006 0.033 1.000000e+00 2
## MOCS3 1.443128e-04 -0.1081431 0.008 0.035 1.000000e+00 2
## HOTAIRM1.1 1.445557e-04 -0.5984325 0.005 0.030 1.000000e+00 2
## AP003717.4.1 1.445636e-04 0.3830094 0.018 0.053 1.000000e+00 2
## NEFL.2 1.449094e-04 -2.5222102 0.003 0.027 1.000000e+00 2
## UIMC1 1.452683e-04 1.1933827 0.122 0.208 1.000000e+00 2
## CLUH.2 1.464120e-04 0.6995051 0.017 0.050 1.000000e+00 2
## ZNF394.1 1.466148e-04 1.0824539 0.137 0.232 1.000000e+00 2
## LY75 1.467995e-04 0.7358299 0.068 0.126 1.000000e+00 2
## PACS2.1 1.468273e-04 1.1727974 0.047 0.095 1.000000e+00 2
## KCTD2 1.470542e-04 0.2812727 0.018 0.053 1.000000e+00 2
## SBNO1-AS1 1.473115e-04 -0.1590852 0.009 0.038 1.000000e+00 2
## CBWD2 1.474119e-04 0.7542674 0.039 0.084 1.000000e+00 2
## OPN3 1.474832e-04 -1.7541664 0.003 0.027 1.000000e+00 2
## PRSS21 1.479118e-04 -0.3439626 0.003 0.027 1.000000e+00 2
## EDC3 1.479594e-04 0.7908396 0.032 0.073 1.000000e+00 2
## AC053513.1 1.480329e-04 0.1805072 0.014 0.045 1.000000e+00 2
## PPIP5K2.2 1.482264e-04 1.2366629 0.149 0.251 1.000000e+00 2
## GPR180.1 1.486218e-04 0.7990221 0.024 0.062 1.000000e+00 2
## WDR35 1.486901e-04 -0.4011770 0.012 0.043 1.000000e+00 2
## ZSCAN26 1.488812e-04 0.1130893 0.021 0.057 1.000000e+00 2
## MAP3K20.2 1.489466e-04 0.5109388 0.009 0.038 1.000000e+00 2
## ZNF792 1.495412e-04 -0.1324758 0.009 0.038 1.000000e+00 2
## C1RL 1.505861e-04 0.9684887 0.063 0.120 1.000000e+00 2
## SEMA4C 1.505871e-04 0.6015317 0.012 0.043 1.000000e+00 2
## CLECL1.1 1.509651e-04 0.7076218 0.014 0.045 1.000000e+00 2
## HEATR1.1 1.509720e-04 1.1750679 0.093 0.164 1.000000e+00 2
## IL17RA 1.510350e-04 1.1549863 0.146 0.245 1.000000e+00 2
## ZBTB10 1.514133e-04 0.8883329 0.012 0.043 1.000000e+00 2
## TCTA.1 1.520204e-04 1.3177868 0.075 0.138 1.000000e+00 2
## RPARP-AS1 1.521124e-04 0.4201410 0.012 0.043 1.000000e+00 2
## QPCTL 1.524560e-04 -0.1591165 0.008 0.035 1.000000e+00 2
## MTOR 1.532794e-04 1.1607666 0.084 0.151 1.000000e+00 2
## KDM6A 1.536087e-04 2.0542614 0.239 0.222 1.000000e+00 2
## FUBP1.1 1.536225e-04 1.1391635 0.209 0.351 1.000000e+00 2
## ZER1 1.541269e-04 0.8332667 0.047 0.095 1.000000e+00 2
## MORC4.1 1.546634e-04 0.3462759 0.005 0.030 1.000000e+00 2
## NR2C1 1.547729e-04 0.9816837 0.069 0.128 1.000000e+00 2
## MANEAL.1 1.551524e-04 -2.1219135 0.002 0.024 1.000000e+00 2
## AC022217.3.1 1.557221e-04 1.4663529 0.119 0.203 1.000000e+00 2
## AP006621.3 1.560042e-04 0.3561384 0.015 0.048 1.000000e+00 2
## COL18A1.1 1.569857e-04 0.6346585 0.015 0.048 1.000000e+00 2
## PRKCZ 1.573012e-04 0.5176816 0.018 0.052 1.000000e+00 2
## C10orf143 1.573284e-04 0.4940582 0.015 0.048 1.000000e+00 2
## GNLY.1 1.576347e-04 -1.8801178 0.033 0.072 1.000000e+00 2
## CALHM6.2 1.582775e-04 -0.9419910 0.003 0.027 1.000000e+00 2
## DLGAP1-AS1 1.583441e-04 1.0353654 0.053 0.104 1.000000e+00 2
## SAV1.2 1.583923e-04 -1.0640946 0.003 0.027 1.000000e+00 2
## CLASP1.1 1.584732e-04 2.0773674 0.236 0.217 1.000000e+00 2
## P2RY14.2 1.586774e-04 -0.1269911 0.011 0.040 1.000000e+00 2
## TRAV19 1.588855e-04 -0.6263604 0.026 0.063 1.000000e+00 2
## EXOC8 1.602119e-04 0.9943415 0.048 0.097 1.000000e+00 2
## CSNK1G3.1 1.602413e-04 1.2292519 0.101 0.176 1.000000e+00 2
## RPP14 1.609983e-04 -0.1621345 0.011 0.040 1.000000e+00 2
## EDRF1.1 1.616593e-04 1.1644256 0.078 0.141 1.000000e+00 2
## LINC01934 1.627255e-04 1.7066777 0.382 0.449 1.000000e+00 2
## CSNK1E 1.636982e-04 1.1919015 0.039 0.084 1.000000e+00 2
## AC016074.2 1.637861e-04 1.3023922 0.039 0.084 1.000000e+00 2
## RASAL3.1 1.644369e-04 1.1431292 0.257 0.437 1.000000e+00 2
## PKD2L2 1.646724e-04 -0.1016374 0.008 0.035 1.000000e+00 2
## DUSP7 1.647629e-04 -1.0878692 0.006 0.032 1.000000e+00 2
## LIG3 1.648991e-04 0.8373558 0.024 0.062 1.000000e+00 2
## BCL7A.1 1.662366e-04 -0.5663188 0.006 0.032 1.000000e+00 2
## SYCE2.2 1.664557e-04 -0.5872584 0.002 0.024 1.000000e+00 2
## CA11 1.664557e-04 -2.0589946 0.002 0.024 1.000000e+00 2
## USP3-AS1 1.671383e-04 0.7801991 0.056 0.108 1.000000e+00 2
## OSBPL2.1 1.671670e-04 0.7646510 0.041 0.086 1.000000e+00 2
## HECTD1 1.672873e-04 1.0119118 0.206 0.344 1.000000e+00 2
## ETS1.1 1.675956e-04 0.5997886 0.397 0.701 1.000000e+00 2
## RBM27 1.676758e-04 1.1603949 0.149 0.249 1.000000e+00 2
## C4orf33.1 1.684785e-04 1.1807839 0.029 0.068 1.000000e+00 2
## PRUNE1 1.687256e-04 0.7161845 0.023 0.059 1.000000e+00 2
## SMARCAL1 1.690527e-04 1.1321705 0.045 0.093 1.000000e+00 2
## RDM1 1.695532e-04 -0.6994632 0.003 0.027 1.000000e+00 2
## HOOK3.1 1.696919e-04 1.1820625 0.152 0.254 1.000000e+00 2
## FANCA.2 1.701162e-04 0.8629018 0.032 0.073 1.000000e+00 2
## WDR60 1.707018e-04 1.3094684 0.071 0.131 1.000000e+00 2
## AGPS.2 1.717425e-04 1.2467991 0.111 0.191 1.000000e+00 2
## ZSCAN25 1.727124e-04 0.6435829 0.036 0.079 1.000000e+00 2
## LYRM9 1.736832e-04 -0.7033507 0.008 0.035 1.000000e+00 2
## MAML2.1 1.737165e-04 1.9715685 0.316 0.336 1.000000e+00 2
## TJAP1 1.742044e-04 0.8970578 0.035 0.077 1.000000e+00 2
## SVIL.2 1.742183e-04 0.9294864 0.033 0.075 1.000000e+00 2
## GGACT 1.748115e-04 -0.2573742 0.009 0.037 1.000000e+00 2
## AC087854.1 1.750797e-04 0.6624215 0.015 0.047 1.000000e+00 2
## TMC8 1.757597e-04 1.1083004 0.268 0.455 1.000000e+00 2
## NCMAP-DT 1.761634e-04 -4.5250586 0.000 0.021 1.000000e+00 2
## LINC01006 1.763113e-04 -1.0508491 0.005 0.030 1.000000e+00 2
## RBMXL1 1.766571e-04 0.3899170 0.015 0.047 1.000000e+00 2
## CARMIL2.1 1.767060e-04 1.4390178 0.089 0.157 1.000000e+00 2
## CERS5.2 1.768211e-04 1.1373141 0.069 0.128 1.000000e+00 2
## SAPCD2.2 1.781723e-04 -1.8185277 0.002 0.024 1.000000e+00 2
## AL022157.1 1.782408e-04 -1.8859549 0.002 0.024 1.000000e+00 2
## DHRS11 1.783778e-04 -1.8357644 0.002 0.024 1.000000e+00 2
## GPATCH1.1 1.785357e-04 0.8232784 0.032 0.073 1.000000e+00 2
## B3GLCT 1.791279e-04 0.5604144 0.020 0.054 1.000000e+00 2
## ZNF431.1 1.798066e-04 1.1062077 0.071 0.130 1.000000e+00 2
## H2AFY2 1.811547e-04 0.2462649 0.011 0.040 1.000000e+00 2
## CTBP1-AS 1.816153e-04 0.2736738 0.009 0.037 1.000000e+00 2
## ARL13B.1 1.816357e-04 0.8149679 0.030 0.070 1.000000e+00 2
## ITGB2-AS1.1 1.835156e-04 1.0278893 0.051 0.101 1.000000e+00 2
## CTLA4.2 1.836051e-04 0.9705829 0.023 0.059 1.000000e+00 2
## C19orf47.1 1.839118e-04 0.5083999 0.012 0.042 1.000000e+00 2
## OSGEPL1 1.839645e-04 0.6615702 0.012 0.042 1.000000e+00 2
## MIR762HG 1.840699e-04 0.7521940 0.012 0.042 1.000000e+00 2
## ZNF780A 1.840755e-04 0.7738845 0.033 0.075 1.000000e+00 2
## ICE1 1.843728e-04 1.2238911 0.138 0.233 1.000000e+00 2
## HPS3 1.844001e-04 1.0016030 0.060 0.114 1.000000e+00 2
## SS18 1.847354e-04 1.0892655 0.152 0.254 1.000000e+00 2
## B3GNTL1.1 1.847827e-04 1.1647429 0.054 0.106 1.000000e+00 2
## SRSF7 1.848663e-04 0.5176835 0.454 0.774 1.000000e+00 2
## TRDMT1.1 1.852484e-04 1.1186999 0.045 0.093 1.000000e+00 2
## NAA30 1.870739e-04 0.8216152 0.036 0.079 1.000000e+00 2
## FAM117B.1 1.871292e-04 0.8808664 0.050 0.099 1.000000e+00 2
## GINS4.2 1.876075e-04 1.2616109 0.018 0.052 1.000000e+00 2
## AC073896.2 1.881773e-04 0.3079490 0.008 0.035 1.000000e+00 2
## ZNF506 1.883156e-04 0.9198740 0.054 0.106 1.000000e+00 2
## CCL5.2 1.885724e-04 -0.4440876 0.656 0.711 1.000000e+00 2
## SNHG21 1.886851e-04 0.6119113 0.015 0.047 1.000000e+00 2
## FAM13B 1.895661e-04 0.9546708 0.162 0.270 1.000000e+00 2
## CTNNA1 1.897796e-04 0.9361175 0.042 0.088 1.000000e+00 2
## CNTRL 1.898119e-04 1.0492037 0.232 0.385 1.000000e+00 2
## RLF 1.899143e-04 1.2462291 0.134 0.226 1.000000e+00 2
## SYT11.1 1.900322e-04 1.1111376 0.080 0.143 1.000000e+00 2
## MDC1.1 1.901663e-04 1.0990357 0.051 0.101 1.000000e+00 2
## ILF3-DT.1 1.909782e-04 1.3693615 0.095 0.165 1.000000e+00 2
## FAM76A 1.918087e-04 0.6641029 0.036 0.079 1.000000e+00 2
## NAA10.1 1.922692e-04 0.7146729 0.026 0.063 1.000000e+00 2
## RBM4B 1.938721e-04 0.6350183 0.021 0.056 1.000000e+00 2
## CEP76.2 1.943004e-04 0.6459743 0.017 0.049 1.000000e+00 2
## WDR53.1 1.947215e-04 1.2329317 0.027 0.066 1.000000e+00 2
## CARM1.1 1.947632e-04 0.9817411 0.017 0.049 1.000000e+00 2
## CYP4V2 1.952264e-04 1.0336886 0.051 0.101 1.000000e+00 2
## WDSUB1.1 1.954767e-04 0.3192170 0.012 0.042 1.000000e+00 2
## SACS.1 1.955090e-04 0.8374443 0.056 0.108 1.000000e+00 2
## B3GNT9 1.959244e-04 0.3701141 0.012 0.042 1.000000e+00 2
## EVL.1 1.972597e-04 0.7020146 0.606 0.839 1.000000e+00 2
## TRAV14DV4.1 1.978749e-04 -0.9560763 0.017 0.049 1.000000e+00 2
## ATF6.1 1.983040e-04 1.0601734 0.144 0.241 1.000000e+00 2
## PLEKHA2 1.991666e-04 1.2424280 0.129 0.218 1.000000e+00 2
## AC079921.1.1 1.997885e-04 0.1874644 0.008 0.034 1.000000e+00 2
## LANCL2.1 2.008354e-04 1.0465495 0.027 0.065 1.000000e+00 2
## YPEL2.1 2.011983e-04 0.8576436 0.057 0.110 1.000000e+00 2
## ANAPC1.1 2.013989e-04 1.0290050 0.069 0.127 1.000000e+00 2
## LRRC23.1 2.015385e-04 1.1797661 0.053 0.103 1.000000e+00 2
## AC009133.3.1 2.019881e-04 -0.2273182 0.009 0.037 1.000000e+00 2
## LIMS2.1 2.026031e-04 -4.6505783 0.000 0.020 1.000000e+00 2
## FAM241B.2 2.026031e-04 -4.0770992 0.000 0.020 1.000000e+00 2
## C5orf34.1 2.030634e-04 0.4279688 0.005 0.029 1.000000e+00 2
## AL096870.8 2.030634e-04 -0.7287444 0.005 0.029 1.000000e+00 2
## TIGD5 2.031876e-04 -0.8361657 0.006 0.032 1.000000e+00 2
## CEP131.1 2.038561e-04 0.1744726 0.006 0.032 1.000000e+00 2
## NUAK2.1 2.041912e-04 -0.8338280 0.006 0.032 1.000000e+00 2
## LINC02482 2.050848e-04 -0.7991513 0.002 0.023 1.000000e+00 2
## MAP3K2-DT 2.050848e-04 -0.9163555 0.002 0.023 1.000000e+00 2
## C5orf22 2.052749e-04 1.2025076 0.032 0.072 1.000000e+00 2
## CCNE2.2 2.057553e-04 1.3290648 0.023 0.059 1.000000e+00 2
## ADCY9.1 2.070998e-04 2.6739719 0.084 0.053 1.000000e+00 2
## GTF2A1 2.071453e-04 1.3598001 0.074 0.134 1.000000e+00 2
## CENPI.2 2.071506e-04 -1.1389647 0.003 0.026 1.000000e+00 2
## ZNF143.1 2.085725e-04 1.4012277 0.072 0.132 1.000000e+00 2
## PNISR.2 2.098933e-04 0.9284837 0.322 0.555 1.000000e+00 2
## ZNF675 2.102900e-04 0.9616727 0.056 0.107 1.000000e+00 2
## APPL2 2.104682e-04 1.1405013 0.068 0.125 1.000000e+00 2
## RPS6KB1 2.112908e-04 1.2846670 0.080 0.143 1.000000e+00 2
## OXSM 2.113448e-04 0.8928983 0.027 0.065 1.000000e+00 2
## SLC4A7 2.120035e-04 1.9940396 0.304 0.319 1.000000e+00 2
## FBXO46 2.121083e-04 0.9787719 0.033 0.074 1.000000e+00 2
## KPNA1 2.121101e-04 1.0714089 0.111 0.189 1.000000e+00 2
## CDR2 2.127801e-04 1.1966564 0.168 0.280 1.000000e+00 2
## RCCD1.2 2.136563e-04 0.8798631 0.021 0.056 1.000000e+00 2
## ZC3H4.1 2.141674e-04 1.2407837 0.057 0.109 1.000000e+00 2
## ZC3H12A 2.142103e-04 1.3110100 0.071 0.129 1.000000e+00 2
## MFSD8 2.149937e-04 0.9225435 0.036 0.079 1.000000e+00 2
## TRAIP.2 2.158343e-04 -0.6673012 0.005 0.029 1.000000e+00 2
## C14orf28 2.169485e-04 -0.7054510 0.005 0.029 1.000000e+00 2
## POLR1B 2.170014e-04 1.1002352 0.039 0.083 1.000000e+00 2
## ATP13A2 2.170038e-04 0.8058987 0.047 0.094 1.000000e+00 2
## TRPC4AP.1 2.176541e-04 1.2200333 0.146 0.243 1.000000e+00 2
## NCOA5.1 2.177587e-04 1.2052818 0.066 0.123 1.000000e+00 2
## ETV7.2 2.198623e-04 -1.4643276 0.002 0.023 1.000000e+00 2
## FUT8 2.199722e-04 1.6946150 0.383 0.461 1.000000e+00 2
## INTS4 2.201587e-04 0.8338729 0.069 0.127 1.000000e+00 2
## PPP1R12A.1 2.201951e-04 0.8957318 0.307 0.529 1.000000e+00 2
## ZRANB3.2 2.203144e-04 0.7863382 0.038 0.081 1.000000e+00 2
## ADAMTSL4-AS1.2 2.208869e-04 2.3073099 0.156 0.123 1.000000e+00 2
## BAHD1 2.221937e-04 0.7197863 0.030 0.069 1.000000e+00 2
## MPRIP 2.222931e-04 1.1891133 0.165 0.275 1.000000e+00 2
## RARA 2.233468e-04 1.3153637 0.099 0.171 1.000000e+00 2
## CEP170.1 2.235274e-04 0.9678221 0.050 0.098 1.000000e+00 2
## DCLRE1B.2 2.249086e-04 0.3472349 0.008 0.034 1.000000e+00 2
## AC021028.1 2.257605e-04 -0.3707807 0.008 0.034 1.000000e+00 2
## SMCHD1 2.260014e-04 1.1947636 0.489 0.679 1.000000e+00 2
## CHD1L.1 2.284782e-04 -0.1748966 0.008 0.034 1.000000e+00 2
## LAX1.1 2.312377e-04 1.0501785 0.066 0.122 1.000000e+00 2
## DUSP18 2.315238e-04 0.7251446 0.020 0.053 1.000000e+00 2
## NUFIP2.1 2.316915e-04 1.8827269 0.334 0.371 1.000000e+00 2
## CHROMR 2.331664e-04 -0.5111347 0.006 0.031 1.000000e+00 2
## URB1-AS1 2.340081e-04 0.6165795 0.006 0.031 1.000000e+00 2
## DCLRE1C.1 2.349963e-04 1.1700526 0.071 0.129 1.000000e+00 2
## CEP95.1 2.351293e-04 1.0248008 0.084 0.149 1.000000e+00 2
## LINC00624 2.353179e-04 -1.7540873 0.003 0.026 1.000000e+00 2
## DNAJC13 2.356098e-04 1.0516679 0.084 0.148 1.000000e+00 2
## LRRC8A 2.358528e-04 1.3774452 0.053 0.103 1.000000e+00 2
## WDR45B 2.358953e-04 1.0901739 0.059 0.111 1.000000e+00 2
## LINC02762 2.361986e-04 0.3866328 0.012 0.042 1.000000e+00 2
## ABHD6 2.364675e-04 0.4976619 0.012 0.042 1.000000e+00 2
## NSUN4 2.369093e-04 0.7572961 0.027 0.065 1.000000e+00 2
## PAQR8.1 2.398589e-04 0.7616571 0.041 0.085 1.000000e+00 2
## ALG9.1 2.399577e-04 1.0483188 0.029 0.067 1.000000e+00 2
## MARCH3.2 2.400845e-04 0.7940489 0.015 0.046 1.000000e+00 2
## SLC39A9.1 2.401528e-04 1.1290527 0.068 0.124 1.000000e+00 2
## GPR174.2 2.414923e-04 1.0638576 0.116 0.196 1.000000e+00 2
## MBTPS2 2.444228e-04 0.6685500 0.015 0.046 1.000000e+00 2
## AC004865.2.1 2.449345e-04 0.2798555 0.008 0.034 1.000000e+00 2
## DCAF17 2.481003e-04 0.9950321 0.035 0.076 1.000000e+00 2
## MAP3K3 2.481804e-04 0.9513788 0.063 0.118 1.000000e+00 2
## MORN3 2.488541e-04 0.9776998 0.014 0.044 1.000000e+00 2
## BTLA 2.496577e-04 0.2652971 0.023 0.058 1.000000e+00 2
## EML2-AS1 2.497680e-04 0.5861155 0.009 0.036 1.000000e+00 2
## ARRB1.1 2.508463e-04 1.2079125 0.059 0.111 1.000000e+00 2
## AL590764.1 2.509482e-04 0.8563348 0.030 0.069 1.000000e+00 2
## ZBTB45 2.521576e-04 1.3157592 0.032 0.071 1.000000e+00 2
## FAM89A 2.526942e-04 -1.2085290 0.002 0.023 1.000000e+00 2
## SPATA24 2.526942e-04 -1.5449249 0.002 0.023 1.000000e+00 2
## SOX12 2.526942e-04 -0.5681774 0.002 0.023 1.000000e+00 2
## PSKH1 2.528027e-04 0.6314969 0.020 0.053 1.000000e+00 2
## TPM1.1 2.533423e-04 0.3302695 0.011 0.039 1.000000e+00 2
## IL23R.2 2.534920e-04 3.5981250 0.026 0.010 1.000000e+00 2
## TMEM150A.1 2.537229e-04 -1.6253946 0.003 0.026 1.000000e+00 2
## S100PBP 2.538046e-04 1.2743242 0.105 0.180 1.000000e+00 2
## METTL8.2 2.543903e-04 0.9439746 0.057 0.108 1.000000e+00 2
## TSPAN15 2.544465e-04 -0.3393364 0.008 0.034 1.000000e+00 2
## HCFC2 2.546658e-04 0.8877151 0.023 0.058 1.000000e+00 2
## SNX8.2 2.555277e-04 1.3034046 0.030 0.069 1.000000e+00 2
## TST 2.562997e-04 -1.0953392 0.003 0.026 1.000000e+00 2
## SCRN1 2.569848e-04 3.5579912 0.014 0.003 1.000000e+00 2
## ASNSD1.1 2.574064e-04 -1.0141658 0.006 0.031 1.000000e+00 2
## KLRG1.2 2.589891e-04 1.0429420 0.138 0.228 1.000000e+00 2
## BCOR.1 2.594672e-04 1.2277460 0.087 0.152 1.000000e+00 2
## TMEM116 2.596657e-04 0.9861253 0.036 0.078 1.000000e+00 2
## FUT11 2.599629e-04 1.0843880 0.036 0.078 1.000000e+00 2
## CC2D1B 2.605541e-04 0.8548557 0.041 0.084 1.000000e+00 2
## KLHL36 2.607755e-04 1.1463737 0.050 0.098 1.000000e+00 2
## RIPOR1 2.621972e-04 0.4587213 0.017 0.048 1.000000e+00 2
## ARNT 2.643208e-04 1.3197018 0.074 0.132 1.000000e+00 2
## TRAF3IP1.1 2.656381e-04 0.9882875 0.033 0.073 1.000000e+00 2
## AKAP10.1 2.667015e-04 1.1393242 0.083 0.146 1.000000e+00 2
## ECD 2.670542e-04 1.2234505 0.137 0.228 1.000000e+00 2
## TMEM91 2.690900e-04 0.4916406 0.021 0.055 1.000000e+00 2
## METAP1D.1 2.699234e-04 0.4041385 0.011 0.039 1.000000e+00 2
## ZNF718.1 2.707002e-04 0.8056544 0.047 0.093 1.000000e+00 2
## DCAF16.1 2.707069e-04 1.0985940 0.054 0.104 1.000000e+00 2
## HPDL.1 2.709086e-04 -0.3140760 0.002 0.023 1.000000e+00 2
## STX18.1 2.715158e-04 1.2091566 0.080 0.141 1.000000e+00 2
## AC007114.1 2.715971e-04 -2.1953530 0.003 0.026 1.000000e+00 2
## VPS33B.1 2.718009e-04 1.1692940 0.026 0.062 1.000000e+00 2
## KNTC1.2 2.720024e-04 1.2075301 0.050 0.097 1.000000e+00 2
## ZNF419 2.731055e-04 -0.2544248 0.008 0.034 1.000000e+00 2
## NADSYN1 2.736794e-04 1.2349287 0.128 0.214 1.000000e+00 2
## RFESD.1 2.737607e-04 -1.0768306 0.003 0.026 1.000000e+00 2
## KIF3B 2.739606e-04 1.1289805 0.051 0.100 1.000000e+00 2
## ZNF451 2.744001e-04 0.9684807 0.108 0.184 1.000000e+00 2
## GPR15.1 2.749323e-04 0.5458704 0.027 0.064 1.000000e+00 2
## FILIP1L.1 2.754201e-04 0.9449383 0.021 0.055 1.000000e+00 2
## ZNF213.1 2.763804e-04 0.2927579 0.008 0.034 1.000000e+00 2
## OPA1 2.778101e-04 1.1663815 0.125 0.208 1.000000e+00 2
## ZNF561 2.793471e-04 0.3534612 0.020 0.053 1.000000e+00 2
## PJA2 2.797996e-04 1.2963233 0.150 0.250 1.000000e+00 2
## TTL 2.799411e-04 0.7674494 0.041 0.084 1.000000e+00 2
## ERN1.1 2.821460e-04 1.5447634 0.368 0.424 1.000000e+00 2
## AC007384.1.1 2.827062e-04 2.3448253 0.179 0.148 1.000000e+00 2
## PLEKHO2 2.833527e-04 0.3356851 0.020 0.053 1.000000e+00 2
## NF2.2 2.836939e-04 0.9808713 0.065 0.119 1.000000e+00 2
## C20orf96.1 2.843287e-04 0.2476203 0.006 0.031 1.000000e+00 2
## CATSPERB 2.848270e-04 2.3551687 0.149 0.116 1.000000e+00 2
## ATP1B1.2 2.850189e-04 1.1503855 0.042 0.086 1.000000e+00 2
## CEP89.1 2.850446e-04 0.9238238 0.024 0.060 1.000000e+00 2
## ZNF8 2.861963e-04 0.4251343 0.017 0.048 1.000000e+00 2
## CERS6.2 2.865907e-04 1.4073088 0.104 0.177 1.000000e+00 2
## MPP7.1 2.867919e-04 1.3677162 0.069 0.126 1.000000e+00 2
## SULT1B1 2.874580e-04 -4.5855838 0.000 0.019 1.000000e+00 2
## ELP3 2.880558e-04 1.1047617 0.099 0.170 1.000000e+00 2
## HACD2 2.882611e-04 0.9528771 0.056 0.106 1.000000e+00 2
## REL.1 2.898725e-04 1.2292768 0.189 0.311 1.000000e+00 2
## PIMREG.2 2.904385e-04 -0.7730555 0.002 0.022 1.000000e+00 2
## AL627171.1 2.904620e-04 0.8978328 0.021 0.055 1.000000e+00 2
## PARN.1 2.935179e-04 1.2878061 0.126 0.211 1.000000e+00 2
## ORC1.2 2.940764e-04 1.1755148 0.017 0.048 1.000000e+00 2
## KLRC2.2 2.945907e-04 -0.1244584 0.008 0.033 1.000000e+00 2
## TRAV38-2DV8 2.947703e-04 -2.0989656 0.012 0.040 1.000000e+00 2
## FCHSD1.1 2.954695e-04 1.2542285 0.051 0.099 1.000000e+00 2
## ZNF134 2.954817e-04 0.3504127 0.021 0.055 1.000000e+00 2
## MICAL1.1 2.967968e-04 1.2004306 0.102 0.174 1.000000e+00 2
## ANKMY2 2.978820e-04 0.5738039 0.032 0.071 1.000000e+00 2
## DUS4L 2.983503e-04 0.6818420 0.014 0.043 1.000000e+00 2
## PPP4R3B 2.993944e-04 1.1417683 0.214 0.356 1.000000e+00 2
## ATL2.1 3.004326e-04 0.7102939 0.053 0.101 1.000000e+00 2
## GNE 3.018368e-04 0.3398726 0.023 0.057 1.000000e+00 2
## G2E3.1 3.018968e-04 1.0912623 0.095 0.163 1.000000e+00 2
## ZNF761 3.038802e-04 0.7643419 0.011 0.038 1.000000e+00 2
## TRGV5.1 3.041053e-04 -0.3674366 0.005 0.028 1.000000e+00 2
## CCDC88A.2 3.043131e-04 0.1099845 0.005 0.028 1.000000e+00 2
## AL365361.1 3.048810e-04 0.4273867 0.015 0.046 1.000000e+00 2
## STX11.1 3.051867e-04 0.7631829 0.044 0.088 1.000000e+00 2
## OGFRL1 3.054656e-04 0.6562929 0.023 0.057 1.000000e+00 2
## RTKN2.2 3.064527e-04 0.3972024 0.014 0.043 1.000000e+00 2
## CASD1 3.089844e-04 1.1254993 0.080 0.141 1.000000e+00 2
## REC8.1 3.118844e-04 0.2612395 0.014 0.043 1.000000e+00 2
## GIGYF1 3.119644e-04 0.7395920 0.038 0.079 1.000000e+00 2
## FAM122C 3.128248e-04 0.8914059 0.062 0.114 1.000000e+00 2
## TBC1D23 3.132568e-04 1.0820126 0.077 0.137 1.000000e+00 2
## KHDC4.1 3.149695e-04 1.0362979 0.050 0.097 1.000000e+00 2
## KAT7 3.158115e-04 1.1358920 0.081 0.143 1.000000e+00 2
## AC044849.1 3.165777e-04 0.8718109 0.057 0.107 1.000000e+00 2
## FOXJ2 3.219827e-04 0.7719003 0.023 0.057 1.000000e+00 2
## THTPA 3.226918e-04 -0.6286973 0.009 0.036 1.000000e+00 2
## ZFX 3.227320e-04 1.0688849 0.096 0.165 1.000000e+00 2
## ATXN7L2 3.241047e-04 0.8447176 0.011 0.038 1.000000e+00 2
## PYCR3 3.247377e-04 0.5779800 0.009 0.036 1.000000e+00 2
## APOM 3.255202e-04 0.6657169 0.009 0.036 1.000000e+00 2
## INPP5E 3.266358e-04 1.1534918 0.021 0.055 1.000000e+00 2
## CTBP1-DT.1 3.283373e-04 0.9472028 0.023 0.057 1.000000e+00 2
## LYSMD3 3.287917e-04 0.7010489 0.062 0.114 1.000000e+00 2
## CEP350 3.293896e-04 0.9922399 0.236 0.391 1.000000e+00 2
## MIR155HG.1 3.305414e-04 1.2395742 0.047 0.092 1.000000e+00 2
## FAM168B 3.306451e-04 1.1484742 0.081 0.143 1.000000e+00 2
## ZNF184 3.312555e-04 0.7804593 0.029 0.066 1.000000e+00 2
## SLC4A2 3.321739e-04 1.1376594 0.036 0.077 1.000000e+00 2
## ELOVL4.1 3.326197e-04 -2.3564486 0.002 0.022 1.000000e+00 2
## RAB3IP.2 3.328802e-04 1.0550155 0.047 0.092 1.000000e+00 2
## DEPDC1.2 3.337052e-04 -0.7602336 0.002 0.022 1.000000e+00 2
## USP45 3.338064e-04 0.8168033 0.032 0.070 1.000000e+00 2
## USP7 3.341702e-04 1.1771633 0.101 0.172 1.000000e+00 2
## NLRP2 3.357600e-04 0.1312090 0.008 0.033 1.000000e+00 2
## AL136419.3.1 3.367493e-04 -0.9918655 0.003 0.025 1.000000e+00 2
## DPYSL2.1 3.370661e-04 1.1039502 0.033 0.073 1.000000e+00 2
## ANTXR2.1 3.376455e-04 1.8500010 0.254 0.245 1.000000e+00 2
## HDAC6 3.385624e-04 0.8961647 0.026 0.061 1.000000e+00 2
## SLC26A6.1 3.396723e-04 1.0741818 0.024 0.059 1.000000e+00 2
## CDKAL1.1 3.403155e-04 1.9289026 0.287 0.295 1.000000e+00 2
## SLC25A15 3.425546e-04 0.6386504 0.009 0.035 1.000000e+00 2
## CYP2R1 3.460114e-04 1.2588373 0.036 0.077 1.000000e+00 2
## ZNF23.1 3.463846e-04 0.1367557 0.009 0.035 1.000000e+00 2
## AC139530.1 3.478723e-04 0.2577348 0.011 0.038 1.000000e+00 2
## CREB3L4 3.478866e-04 -0.6584619 0.006 0.030 1.000000e+00 2
## RAC3.2 3.482261e-04 0.5151003 0.006 0.030 1.000000e+00 2
## FAM102B.2 3.487913e-04 0.9585039 0.045 0.090 1.000000e+00 2
## MIEF2 3.493151e-04 0.4602848 0.005 0.028 1.000000e+00 2
## ZNF473.2 3.520637e-04 0.6679059 0.018 0.050 1.000000e+00 2
## PARP14.2 3.534104e-04 1.1779630 0.208 0.340 1.000000e+00 2
## SLC25A16.1 3.543496e-04 1.0671346 0.047 0.092 1.000000e+00 2
## MMEL1.1 3.546605e-04 -4.5134257 0.000 0.019 1.000000e+00 2
## AL139246.5.1 3.549042e-04 1.2736611 0.029 0.066 1.000000e+00 2
## ATP1A1-AS1 3.553771e-04 0.5005298 0.014 0.043 1.000000e+00 2
## ALKBH1 3.616119e-04 0.7466430 0.023 0.057 1.000000e+00 2
## RPAP2 3.626376e-04 1.3409490 0.167 0.274 1.000000e+00 2
## FYN.2 3.660192e-04 1.6095679 0.408 0.511 1.000000e+00 2
## FAM160A2.1 3.681522e-04 1.3089858 0.044 0.087 1.000000e+00 2
## GALNT6.1 3.691183e-04 1.3535816 0.041 0.083 1.000000e+00 2
## PFKM.1 3.693479e-04 0.1939555 0.009 0.035 1.000000e+00 2
## ZNF717 3.698176e-04 -0.6385866 0.006 0.030 1.000000e+00 2
## CNBD2 3.710149e-04 0.7262253 0.015 0.045 1.000000e+00 2
## SDE2 3.717050e-04 0.9855540 0.056 0.105 1.000000e+00 2
## DENND11 3.720399e-04 1.0360725 0.057 0.107 1.000000e+00 2
## CHRM3 3.722747e-04 -0.7612968 0.005 0.027 1.000000e+00 2
## EIF4ENIF1 3.728313e-04 1.0547730 0.048 0.094 1.000000e+00 2
## DNAL1 3.729088e-04 -0.4298671 0.005 0.027 1.000000e+00 2
## AIFM2 3.732898e-04 -0.7411304 0.005 0.027 1.000000e+00 2
## ARNTL2.2 3.733201e-04 0.6913815 0.026 0.061 1.000000e+00 2
## CCZ1B 3.734168e-04 -0.4015810 0.005 0.027 1.000000e+00 2
## ZNF331.1 3.764868e-04 1.4021281 0.159 0.258 1.000000e+00 2
## PHF20 3.767210e-04 1.1851747 0.202 0.329 1.000000e+00 2
## MSH5.1 3.774284e-04 0.7404953 0.021 0.054 1.000000e+00 2
## AC006504.5 3.780893e-04 -0.1457984 0.012 0.040 1.000000e+00 2
## DPY19L1.1 3.791585e-04 0.8898387 0.053 0.100 1.000000e+00 2
## KHDC1.1 3.793082e-04 0.8015057 0.018 0.050 1.000000e+00 2
## AC025159.1 3.797771e-04 1.0569218 0.027 0.063 1.000000e+00 2
## FAM193A.1 3.798810e-04 1.1973773 0.098 0.166 1.000000e+00 2
## PREP 3.818741e-04 1.0921752 0.107 0.180 1.000000e+00 2
## NUDT19 3.825243e-04 0.9119240 0.032 0.070 1.000000e+00 2
## SKIV2L 3.843905e-04 1.2894152 0.053 0.101 1.000000e+00 2
## GPR89A 3.852704e-04 0.9729479 0.035 0.074 1.000000e+00 2
## GALM.1 3.860202e-04 1.3704140 0.134 0.221 1.000000e+00 2
## NCK1 3.863096e-04 1.2628124 0.128 0.213 1.000000e+00 2
## C14orf93 3.863978e-04 0.6900001 0.015 0.045 1.000000e+00 2
## TMEM30A.1 3.879035e-04 0.9463133 0.162 0.263 1.000000e+00 2
## RMND5B.2 3.896806e-04 1.3229816 0.038 0.079 1.000000e+00 2
## ALG14.1 3.903107e-04 0.6671421 0.015 0.045 1.000000e+00 2
## RBM41 3.921167e-04 0.8547086 0.039 0.081 1.000000e+00 2
## UHRF1BP1L 3.925571e-04 0.9975326 0.087 0.151 1.000000e+00 2
## ATG13.1 3.962734e-04 1.1771977 0.096 0.164 1.000000e+00 2
## FBXW7.1 3.971884e-04 1.8430806 0.322 0.353 1.000000e+00 2
## NR1H3 3.976865e-04 -0.2211450 0.005 0.027 1.000000e+00 2
## TRBV7-6.1 3.986266e-04 -2.8009549 0.005 0.027 1.000000e+00 2
## AC090152.1.2 3.989054e-04 -0.8459192 0.005 0.027 1.000000e+00 2
## ATG9A 3.994801e-04 0.9618454 0.029 0.065 1.000000e+00 2
## SLC7A6 3.997338e-04 1.2851870 0.120 0.199 1.000000e+00 2
## SPATA2 3.999613e-04 -0.5205238 0.012 0.040 1.000000e+00 2
## CCDC50 4.001143e-04 -0.1400117 0.008 0.032 1.000000e+00 2
## TM6SF1.1 4.017592e-04 0.5706132 0.018 0.049 1.000000e+00 2
## RAB28.1 4.077233e-04 1.3091549 0.116 0.193 1.000000e+00 2
## KMO 4.080161e-04 -4.3208378 0.000 0.018 1.000000e+00 2
## ZNF92 4.109153e-04 1.0685314 0.087 0.150 1.000000e+00 2
## PIP4K2B 4.117105e-04 0.8935371 0.038 0.079 1.000000e+00 2
## CCL4L2.2 4.124414e-04 -1.9684254 0.003 0.024 1.000000e+00 2
## TRAF5.1 4.125398e-04 1.0397109 0.165 0.271 1.000000e+00 2
## AGFG1.1 4.146645e-04 1.2885937 0.132 0.219 1.000000e+00 2
## GATD1 4.152092e-04 1.2267100 0.036 0.076 1.000000e+00 2
## MDM2 4.168841e-04 1.1194555 0.174 0.284 1.000000e+00 2
## PROSER1 4.232697e-04 0.8560199 0.060 0.111 1.000000e+00 2
## FOSL2.1 4.248058e-04 1.4828703 0.056 0.104 1.000000e+00 2
## DHODH 4.269336e-04 0.2222243 0.014 0.042 1.000000e+00 2
## ZNRF1.1 4.270126e-04 0.5078556 0.024 0.058 1.000000e+00 2
## ERLIN1.1 4.276450e-04 0.8746280 0.026 0.060 1.000000e+00 2
## CHKA 4.278729e-04 0.5171908 0.018 0.049 1.000000e+00 2
## AL606489.1 4.286288e-04 -0.6506292 0.011 0.037 1.000000e+00 2
## ATP13A3.1 4.319989e-04 1.0634158 0.093 0.159 1.000000e+00 2
## HAUS5.2 4.325612e-04 1.4540046 0.051 0.098 1.000000e+00 2
## HIST1H2AB.1 4.340995e-04 1.0077590 0.015 0.044 1.000000e+00 2
## NOL10.1 4.349598e-04 1.2801066 0.084 0.146 1.000000e+00 2
## ZNF821 4.385979e-04 1.1413579 0.042 0.085 1.000000e+00 2
## EPOR 4.392902e-04 0.6891351 0.015 0.044 1.000000e+00 2
## NCF2 4.395844e-04 -0.5348009 0.008 0.032 1.000000e+00 2
## LINC00526 4.397498e-04 -2.5311170 0.002 0.021 1.000000e+00 2
## MLH3 4.400057e-04 0.7335550 0.033 0.071 1.000000e+00 2
## AC211476.2 4.409299e-04 -0.8299132 0.002 0.021 1.000000e+00 2
## FKBP1C 4.409299e-04 -1.4548167 0.002 0.021 1.000000e+00 2
## CMIP 4.425262e-04 2.0862229 0.236 0.222 1.000000e+00 2
## PDE5A 4.429724e-04 -0.2203668 0.011 0.037 1.000000e+00 2
## HILPDA.1 4.433637e-04 -0.2961818 0.006 0.030 1.000000e+00 2
## MPP6.1 4.448904e-04 1.4352506 0.039 0.080 1.000000e+00 2
## CPNE2 4.468680e-04 0.5433762 0.009 0.035 1.000000e+00 2
## XYLB.2 4.472074e-04 1.1836881 0.011 0.037 1.000000e+00 2
## KIAA0232.1 4.491215e-04 1.2049213 0.089 0.152 1.000000e+00 2
## UTP25 4.496470e-04 0.7709831 0.026 0.060 1.000000e+00 2
## BRPF1 4.497307e-04 1.2424404 0.056 0.104 1.000000e+00 2
## MTMR1 4.500607e-04 1.1952939 0.090 0.154 1.000000e+00 2
## SHLD2.1 4.504958e-04 1.1242498 0.113 0.188 1.000000e+00 2
## ZNF699 4.520505e-04 0.6403524 0.024 0.058 1.000000e+00 2
## NAP1L5 4.524969e-04 -0.1058126 0.006 0.030 1.000000e+00 2
## DHX32 4.526281e-04 1.0177067 0.036 0.076 1.000000e+00 2
## AC026979.2 4.527463e-04 0.9381326 0.017 0.047 1.000000e+00 2
## COQ7 4.554868e-04 1.6299873 0.072 0.128 1.000000e+00 2
## GSTCD.2 4.555285e-04 0.9589921 0.018 0.049 1.000000e+00 2
## MPHOSPH8 4.562589e-04 1.1136990 0.265 0.443 1.000000e+00 2
## AC090204.1.1 4.578387e-04 0.5318569 0.005 0.027 1.000000e+00 2
## RNF185 4.604342e-04 1.0182672 0.036 0.076 1.000000e+00 2
## NSMAF.1 4.609237e-04 1.2910220 0.120 0.200 1.000000e+00 2
## CNOT6 4.611640e-04 1.1635465 0.063 0.115 1.000000e+00 2
## TMED8.1 4.619600e-04 1.2755639 0.042 0.085 1.000000e+00 2
## BIVM 4.637726e-04 1.1071071 0.041 0.082 1.000000e+00 2
## WRN.2 4.654741e-04 1.2724621 0.074 0.131 1.000000e+00 2
## TRBV5-1 4.662633e-04 -0.9270077 0.021 0.052 1.000000e+00 2
## GIN1 4.664885e-04 0.5237232 0.015 0.044 1.000000e+00 2
## MLKL.1 4.667471e-04 1.2085841 0.047 0.091 1.000000e+00 2
## CCDC84-DT.2 4.685846e-04 0.4705331 0.008 0.032 1.000000e+00 2
## ZNF182 4.694358e-04 -4.2281525 0.000 0.018 1.000000e+00 2
## BEX5 4.694358e-04 -4.2745715 0.000 0.018 1.000000e+00 2
## SPEF2 4.714479e-04 0.8878939 0.024 0.058 1.000000e+00 2
## CCDC200.1 4.722294e-04 0.7367506 0.039 0.080 1.000000e+00 2
## AC021683.5 4.738097e-04 0.5318349 0.011 0.037 1.000000e+00 2
## ZNF513 4.755464e-04 0.4430882 0.018 0.049 1.000000e+00 2
## HDAC5.1 4.760383e-04 0.7826240 0.026 0.060 1.000000e+00 2
## DUSP4 4.778509e-04 1.0983311 0.050 0.095 1.000000e+00 2
## TSHZ2.2 4.788513e-04 1.2617511 0.054 0.102 1.000000e+00 2
## NIN.1 4.796960e-04 1.0268064 0.251 0.413 1.000000e+00 2
## AMD1 4.840175e-04 1.4550812 0.143 0.235 1.000000e+00 2
## HNRNPLL.2 4.841816e-04 1.1697916 0.150 0.246 1.000000e+00 2
## PRPF4B 4.863513e-04 1.1405318 0.253 0.420 1.000000e+00 2
## BOLA2B 4.881039e-04 0.8195058 0.014 0.042 1.000000e+00 2
## PTDSS2.1 4.889369e-04 0.9034857 0.023 0.056 1.000000e+00 2
## CNGA4 4.908128e-04 3.4338846 0.017 0.005 1.000000e+00 2
## SLC7A1.2 4.913586e-04 1.2493683 0.078 0.136 1.000000e+00 2
## ZXDC 4.944413e-04 1.1170668 0.078 0.136 1.000000e+00 2
## SENP1.1 4.966275e-04 1.2697765 0.074 0.130 1.000000e+00 2
## RAB43.1 4.978305e-04 1.1133324 0.036 0.075 1.000000e+00 2
## RC3H2.1 4.988663e-04 1.2936741 0.116 0.192 1.000000e+00 2
## APOBEC3B.2 5.035463e-04 -3.9437525 0.000 0.018 1.000000e+00 2
## ABHD15-AS1 5.036911e-04 0.3800159 0.018 0.049 1.000000e+00 2
## PDPR 5.067980e-04 0.8575104 0.048 0.093 1.000000e+00 2
## SORD 5.068796e-04 -1.6144557 0.002 0.021 1.000000e+00 2
## AC018521.5 5.069571e-04 0.9237102 0.021 0.053 1.000000e+00 2
## ZNF345 5.073422e-04 0.3198504 0.011 0.037 1.000000e+00 2
## AP002490.1 5.141466e-04 0.8408642 0.014 0.042 1.000000e+00 2
## ZBP1.1 5.172470e-04 1.0591686 0.042 0.084 1.000000e+00 2
## ACTL10 5.181605e-04 -0.6159419 0.006 0.029 1.000000e+00 2
## PCBD2.2 5.192798e-04 1.1734822 0.054 0.101 1.000000e+00 2
## FAM214B 5.202953e-04 -0.3791326 0.012 0.039 1.000000e+00 2
## KMT5B 5.209817e-04 1.1004319 0.099 0.167 1.000000e+00 2
## TRBJ2-3 5.220751e-04 -1.0501688 0.005 0.026 1.000000e+00 2
## LDAH 5.239913e-04 1.1784225 0.089 0.152 1.000000e+00 2
## PIGM 5.252172e-04 0.4738222 0.024 0.058 1.000000e+00 2
## MYO19.2 5.274439e-04 1.2083939 0.068 0.120 1.000000e+00 2
## SPG7.1 5.295723e-04 1.4125816 0.141 0.231 1.000000e+00 2
## ZNF681.1 5.330586e-04 -0.1122818 0.008 0.032 1.000000e+00 2
## RINT1 5.336684e-04 0.9455819 0.039 0.079 1.000000e+00 2
## PPP2R5B 5.340346e-04 0.9486316 0.023 0.055 1.000000e+00 2
## PTPRA 5.347923e-04 1.1343918 0.203 0.333 1.000000e+00 2
## UBE2H 5.355666e-04 1.1568942 0.198 0.325 1.000000e+00 2
## ZNF846 5.373623e-04 0.9555532 0.017 0.046 1.000000e+00 2
## NOP9 5.405242e-04 0.9406482 0.020 0.051 1.000000e+00 2
## C2orf27A 5.426093e-04 0.7823185 0.024 0.057 1.000000e+00 2
## NRAV 5.436852e-04 -1.5180935 0.002 0.021 1.000000e+00 2
## SCAF4.1 5.438538e-04 1.1872050 0.131 0.215 1.000000e+00 2
## SWAP70.1 5.444616e-04 0.7421252 0.027 0.062 1.000000e+00 2
## SLC25A29 5.465415e-04 0.6340281 0.023 0.055 1.000000e+00 2
## LARP1B.2 5.496328e-04 1.3505110 0.075 0.132 1.000000e+00 2
## MAPK11 5.505356e-04 1.0096629 0.014 0.041 1.000000e+00 2
## TLE3 5.507917e-04 0.8871343 0.038 0.077 1.000000e+00 2
## ACAP1.1 5.546435e-04 0.8983818 0.374 0.647 1.000000e+00 2
## SYNGAP1 5.565405e-04 1.3068838 0.045 0.088 1.000000e+00 2
## COG7 5.581529e-04 1.4111252 0.051 0.097 1.000000e+00 2
## HMG20A.1 5.586383e-04 1.0864499 0.081 0.140 1.000000e+00 2
## MBTPS1 5.588432e-04 1.1961848 0.110 0.183 1.000000e+00 2
## KLHL21 5.591427e-04 1.1015886 0.024 0.057 1.000000e+00 2
## CLIP1.1 5.593646e-04 1.1583210 0.185 0.300 1.000000e+00 2
## PRMT3.2 5.618573e-04 1.1935051 0.074 0.129 1.000000e+00 2
## ZNF17 5.629381e-04 -0.6198226 0.008 0.031 1.000000e+00 2
## ZNF398 5.634833e-04 1.1322494 0.051 0.096 1.000000e+00 2
## GALNT12.2 5.640205e-04 0.2209988 0.012 0.039 1.000000e+00 2
## PKD2.1 5.641028e-04 1.0164088 0.060 0.109 1.000000e+00 2
## AC245407.2.1 5.641235e-04 0.9328179 0.015 0.044 1.000000e+00 2
## UBE3B.1 5.645052e-04 1.0615836 0.036 0.075 1.000000e+00 2
## FAM160B1 5.650484e-04 1.2294531 0.078 0.136 1.000000e+00 2
## ZBTB9 5.651161e-04 -0.4318726 0.009 0.034 1.000000e+00 2
## GFOD1.2 5.656820e-04 3.1602114 0.036 0.017 1.000000e+00 2
## HIRA 5.680771e-04 0.4404268 0.020 0.051 1.000000e+00 2
## NMNAT1 5.709798e-04 0.1151499 0.008 0.031 1.000000e+00 2
## CHST7.1 5.716714e-04 0.8672767 0.026 0.059 1.000000e+00 2
## BFSP2.1 5.751958e-04 -2.9729923 0.002 0.021 1.000000e+00 2
## TOGARAM1 5.767043e-04 1.0692779 0.060 0.109 1.000000e+00 2
## ABCD1.1 5.786137e-04 0.6381452 0.014 0.041 1.000000e+00 2
## ZNF575 5.788554e-04 0.1590063 0.009 0.034 1.000000e+00 2
## AC100861.1 5.794223e-04 -4.2087853 0.000 0.018 1.000000e+00 2
## SPHK1 5.794223e-04 -3.9738652 0.000 0.018 1.000000e+00 2
## SMTN.1 5.794223e-04 -4.3445280 0.000 0.018 1.000000e+00 2
## FAM72B.2 5.798359e-04 -2.5243925 0.002 0.021 1.000000e+00 2
## CCL4.2 5.801995e-04 -0.1236452 0.029 0.063 1.000000e+00 2
## ZSCAN2 5.810892e-04 0.5135721 0.009 0.034 1.000000e+00 2
## SENCR 5.829486e-04 -0.7893181 0.002 0.021 1.000000e+00 2
## TAPT1.1 5.831084e-04 1.2430142 0.060 0.110 1.000000e+00 2
## KBTBD7 5.843975e-04 0.5137591 0.014 0.041 1.000000e+00 2
## NRGN.2 5.854800e-04 0.1099051 0.003 0.023 1.000000e+00 2
## AC004556.3.1 5.854800e-04 -0.3625245 0.003 0.023 1.000000e+00 2
## TCAF1.1 5.874427e-04 0.4078399 0.023 0.055 1.000000e+00 2
## SESN2.1 5.904004e-04 1.1668260 0.047 0.090 1.000000e+00 2
## ECPAS 5.918491e-04 1.2975013 0.122 0.200 1.000000e+00 2
## USP18.1 5.942362e-04 0.5376636 0.006 0.029 1.000000e+00 2
## KLF11.1 5.962976e-04 0.1746361 0.012 0.039 1.000000e+00 2
## SP140L.1 5.963130e-04 1.1388937 0.245 0.406 1.000000e+00 2
## DPY19L4 5.966093e-04 0.6558532 0.017 0.046 1.000000e+00 2
## C5orf30.1 5.975165e-04 -0.4998241 0.005 0.026 1.000000e+00 2
## SUSD6.1 5.979584e-04 2.3017723 0.177 0.150 1.000000e+00 2
## MEF2D.1 5.983991e-04 1.3472947 0.066 0.118 1.000000e+00 2
## RETSAT 5.991967e-04 0.9322357 0.032 0.068 1.000000e+00 2
## ENTPD1-AS1.1 5.993471e-04 1.0399512 0.063 0.114 1.000000e+00 2
## P3H1 5.996260e-04 1.3476383 0.041 0.081 1.000000e+00 2
## VIPR1.1 6.003476e-04 -0.6145362 0.005 0.026 1.000000e+00 2
## DYNC2LI1 6.007693e-04 0.2341127 0.012 0.039 1.000000e+00 2
## DFFB 6.016008e-04 0.2289059 0.012 0.039 1.000000e+00 2
## VAC14.1 6.025861e-04 1.2851519 0.045 0.088 1.000000e+00 2
## INO80D 6.062093e-04 1.2462263 0.122 0.200 1.000000e+00 2
## ZNF507 6.062721e-04 1.2581435 0.059 0.107 1.000000e+00 2
## CREBRF 6.065162e-04 1.0489982 0.098 0.164 1.000000e+00 2
## ANKRD10 6.077753e-04 1.1287008 0.161 0.262 1.000000e+00 2
## ALKBH8 6.110374e-04 0.7640304 0.018 0.048 1.000000e+00 2
## SP1.1 6.144343e-04 1.2252173 0.044 0.086 1.000000e+00 2
## NCK1-DT 6.149916e-04 1.0183144 0.018 0.048 1.000000e+00 2
## ASRGL1.2 6.165108e-04 -3.0896951 0.002 0.020 1.000000e+00 2
## DHDDS.1 6.165250e-04 1.0277103 0.047 0.090 1.000000e+00 2
## ATG4A 6.188011e-04 0.7554125 0.021 0.053 1.000000e+00 2
## SLC25A35 6.229731e-04 0.3945238 0.014 0.041 1.000000e+00 2
## BCL2L11.1 6.233138e-04 0.3824539 0.035 0.072 1.000000e+00 2
## LACTB2-AS1 6.245793e-04 -1.9647517 0.002 0.020 1.000000e+00 2
## PTPRO.1 6.250570e-04 -0.7277669 0.002 0.020 1.000000e+00 2
## RAB11B-AS1 6.264915e-04 -0.6243717 0.003 0.023 1.000000e+00 2
## NADK2 6.286016e-04 0.5116932 0.020 0.050 1.000000e+00 2
## CAD.1 6.331946e-04 0.9921164 0.036 0.074 1.000000e+00 2
## E2F7.2 6.354665e-04 0.6102298 0.006 0.029 1.000000e+00 2
## DPH6.2 6.361594e-04 1.4687881 0.068 0.120 1.000000e+00 2
## STAM2 6.362447e-04 1.1888584 0.057 0.105 1.000000e+00 2
## LINC01393 6.364620e-04 2.5336289 0.074 0.046 1.000000e+00 2
## CARNMT1.1 6.392575e-04 1.1272142 0.084 0.144 1.000000e+00 2
## PUS7.2 6.406706e-04 1.3924943 0.023 0.055 1.000000e+00 2
## C16orf95.2 6.419948e-04 0.9952389 0.012 0.038 1.000000e+00 2
## ARHGEF19 6.423166e-04 0.6949545 0.017 0.046 1.000000e+00 2
## ENTPD5.1 6.423492e-04 0.8991570 0.012 0.038 1.000000e+00 2
## ROPN1L 6.423518e-04 0.2173049 0.005 0.026 1.000000e+00 2
## C17orf53.2 6.446299e-04 0.1986154 0.008 0.031 1.000000e+00 2
## KDM2B 6.478268e-04 1.3299784 0.119 0.196 1.000000e+00 2
## CROT 6.480299e-04 0.6831355 0.030 0.066 1.000000e+00 2
## ABTB2.1 6.485025e-04 2.6534953 0.077 0.049 1.000000e+00 2
## CLIC5.2 6.492286e-04 0.8298911 0.032 0.068 1.000000e+00 2
## CKAP4.1 6.543668e-04 0.5354540 0.009 0.034 1.000000e+00 2
## ATF2 6.544034e-04 1.1785270 0.117 0.193 1.000000e+00 2
## RNF170 6.549629e-04 1.0243855 0.036 0.074 1.000000e+00 2
## OSGIN2 6.556475e-04 1.2212803 0.032 0.068 1.000000e+00 2
## CENPC 6.601702e-04 1.3015430 0.202 0.330 1.000000e+00 2
## ZKSCAN8 6.610520e-04 0.6987639 0.024 0.057 1.000000e+00 2
## AC104365.1 6.648773e-04 -0.2094735 0.006 0.028 1.000000e+00 2
## AUNIP.2 6.667955e-04 -3.6197889 0.000 0.017 1.000000e+00 2
## C2orf69 6.677689e-04 0.6042822 0.024 0.057 1.000000e+00 2
## MUC1 6.699615e-04 -2.0153668 0.002 0.020 1.000000e+00 2
## SDC4.1 6.706678e-04 0.7604440 0.012 0.038 1.000000e+00 2
## ETV2 6.718142e-04 -0.1825474 0.003 0.023 1.000000e+00 2
## NFIL3 6.746661e-04 1.0508167 0.084 0.144 1.000000e+00 2
## AC099063.1.1 6.803501e-04 0.3423714 0.012 0.038 1.000000e+00 2
## AC005837.1 6.823723e-04 3.7117197 0.014 0.004 1.000000e+00 2
## NT5M 6.824680e-04 0.6430044 0.020 0.050 1.000000e+00 2
## ACSS2.2 6.834575e-04 0.7684926 0.020 0.050 1.000000e+00 2
## NSUN3 6.848242e-04 1.0508403 0.048 0.092 1.000000e+00 2
## SLC35B4 6.858481e-04 0.8501089 0.030 0.066 1.000000e+00 2
## ZNF273.2 6.864264e-04 1.2028908 0.021 0.052 1.000000e+00 2
## AC103563.7 6.870538e-04 -0.7756837 0.005 0.026 1.000000e+00 2
## WDCP 6.916952e-04 0.4401330 0.014 0.041 1.000000e+00 2
## PTER 6.933575e-04 1.2227540 0.035 0.072 1.000000e+00 2
## TMEM201.2 6.957208e-04 0.3191543 0.008 0.031 1.000000e+00 2
## PIGB 6.996352e-04 0.9905334 0.054 0.100 1.000000e+00 2
## PILRA 7.022179e-04 3.7451613 0.021 0.008 1.000000e+00 2
## CPPED1.1 7.024724e-04 1.0618870 0.045 0.087 1.000000e+00 2
## SBF1 7.055602e-04 1.3011387 0.077 0.132 1.000000e+00 2
## PLCL1.2 7.088346e-04 1.1906169 0.050 0.093 1.000000e+00 2
## SPOPL.1 7.140473e-04 1.0341702 0.039 0.078 1.000000e+00 2
## AGPAT4.2 7.143908e-04 1.0315008 0.075 0.130 1.000000e+00 2
## TAB1 7.153008e-04 1.2539525 0.044 0.085 1.000000e+00 2
## PRXL2A.2 7.153327e-04 -3.9275645 0.000 0.017 1.000000e+00 2
## TRBV11-2 7.162603e-04 -2.1100963 0.006 0.028 1.000000e+00 2
## ZNF621 7.163222e-04 0.9243378 0.027 0.061 1.000000e+00 2
## IFNL1.1 7.186529e-04 -1.6872917 0.002 0.020 1.000000e+00 2
## BFSP1.1 7.191319e-04 -0.4348946 0.003 0.023 1.000000e+00 2
## INSL6 7.191810e-04 0.5935286 0.018 0.047 1.000000e+00 2
## TECPR1 7.206128e-04 1.1554682 0.080 0.136 1.000000e+00 2
## EPS15L1 7.210810e-04 1.3313140 0.101 0.168 1.000000e+00 2
## IVD.1 7.234585e-04 1.1800645 0.086 0.146 1.000000e+00 2
## ANKRD16.1 7.251391e-04 0.8359813 0.012 0.038 1.000000e+00 2
## SDHAF4 7.261246e-04 0.6325225 0.020 0.050 1.000000e+00 2
## PBRM1.1 7.335216e-04 1.1094155 0.217 0.352 1.000000e+00 2
## PITRM1.1 7.343349e-04 1.3858501 0.068 0.120 1.000000e+00 2
## NEK2.2 7.343624e-04 0.6472575 0.005 0.026 1.000000e+00 2
## EXO5 7.348566e-04 0.4239151 0.005 0.026 1.000000e+00 2
## DDI2 7.362076e-04 0.9648929 0.039 0.078 1.000000e+00 2
## ZMAT3.1 7.398230e-04 0.8882528 0.116 0.188 1.000000e+00 2
## HIC2.1 7.425699e-04 0.6150924 0.011 0.036 1.000000e+00 2
## CCDC126 7.429933e-04 0.8007013 0.023 0.054 1.000000e+00 2
## ANKAR 7.455603e-04 1.0198672 0.042 0.083 1.000000e+00 2
## ACER3.1 7.465722e-04 1.0303226 0.035 0.072 1.000000e+00 2
## RUNX2.2 7.494179e-04 2.3137034 0.208 0.188 1.000000e+00 2
## ERCC6L2 7.497350e-04 1.1767519 0.123 0.201 1.000000e+00 2
## ZNF525 7.523065e-04 0.4514672 0.012 0.038 1.000000e+00 2
## SDCBP2-AS1 7.539052e-04 0.9835351 0.029 0.063 1.000000e+00 2
## NOL9 7.620589e-04 1.3241557 0.087 0.148 1.000000e+00 2
## ATG14.1 7.644867e-04 1.0080890 0.050 0.093 1.000000e+00 2
## NHLRC4 7.651352e-04 -1.5187117 0.003 0.023 1.000000e+00 2
## AC008440.1 7.657403e-04 0.9215273 0.026 0.059 1.000000e+00 2
## METTL14 7.666376e-04 1.1705558 0.051 0.096 1.000000e+00 2
## FAM13A.1 7.674158e-04 0.7288745 0.032 0.067 1.000000e+00 2
## SHPK.1 7.678438e-04 0.9347941 0.017 0.045 1.000000e+00 2
## PTPN1.1 7.723836e-04 1.2055692 0.265 0.440 1.000000e+00 2
## ZSCAN29 7.782640e-04 -0.1179648 0.011 0.035 1.000000e+00 2
## IDI1.1 7.787075e-04 1.2181592 0.174 0.281 1.000000e+00 2
## PDS5A 7.797173e-04 1.0280601 0.250 0.411 1.000000e+00 2
## RNF123 7.814384e-04 0.5182219 0.021 0.052 1.000000e+00 2
## SYCE1L 7.836018e-04 -0.1342418 0.005 0.025 1.000000e+00 2
## SCAMP1 7.849486e-04 1.2849125 0.068 0.119 1.000000e+00 2
## PGGT1B.1 7.869898e-04 0.9534350 0.054 0.099 1.000000e+00 2
## KMT2E-AS1 7.888132e-04 0.6965755 0.027 0.061 1.000000e+00 2
## FBXO31 7.891750e-04 1.2219321 0.059 0.106 1.000000e+00 2
## DGCR2 7.895104e-04 0.9014462 0.068 0.119 1.000000e+00 2
## PARP16.2 7.895630e-04 1.2353503 0.048 0.091 1.000000e+00 2
## ARMC7 7.904341e-04 0.4229308 0.018 0.047 1.000000e+00 2
## LZTR1 7.910858e-04 0.7662629 0.030 0.065 1.000000e+00 2
## TBC1D31.1 7.911400e-04 1.3942878 0.065 0.115 1.000000e+00 2
## KIAA0100.1 7.933089e-04 1.2444680 0.111 0.183 1.000000e+00 2
## PPP1R3D 7.957737e-04 0.7990438 0.014 0.040 1.000000e+00 2
## HIPK3 7.991714e-04 1.2658934 0.105 0.174 1.000000e+00 2
## RPAP1 8.026962e-04 0.8103587 0.026 0.058 1.000000e+00 2
## HAVCR2.1 8.027491e-04 1.2839385 0.065 0.114 1.000000e+00 2
## SLC25A53 8.081296e-04 0.6929979 0.029 0.063 1.000000e+00 2
## ETS2 8.085633e-04 1.0849948 0.032 0.067 1.000000e+00 2
## ASH1L-AS1 8.096188e-04 0.2785970 0.017 0.045 1.000000e+00 2
## ZNF133 8.125958e-04 0.9486106 0.015 0.043 1.000000e+00 2
## PCNX4 8.149748e-04 1.1966995 0.126 0.204 1.000000e+00 2
## TRIM41.1 8.164230e-04 1.2572216 0.057 0.104 1.000000e+00 2
## CTNNB1 8.166288e-04 1.1258457 0.176 0.285 1.000000e+00 2
## SLC37A3 8.202936e-04 1.2609459 0.044 0.084 1.000000e+00 2
## TP53I3 8.233309e-04 -4.1310566 0.000 0.017 1.000000e+00 2
## ZNF530.1 8.237127e-04 -0.8567421 0.003 0.022 1.000000e+00 2
## DCAKD.1 8.240514e-04 1.3841669 0.050 0.093 1.000000e+00 2
## SH2D3C.1 8.243362e-04 1.3215830 0.117 0.191 1.000000e+00 2
## PCSK1N.1 8.244300e-04 -2.2490346 0.002 0.020 1.000000e+00 2
## AK1 8.248229e-04 2.9968749 0.051 0.029 1.000000e+00 2
## AL137857.1 8.256900e-04 -1.9964689 0.002 0.020 1.000000e+00 2
## MTREX 8.269494e-04 1.2639556 0.134 0.217 1.000000e+00 2
## CLN8 8.303967e-04 0.4921574 0.014 0.040 1.000000e+00 2
## WARS2 8.304394e-04 1.4208190 0.102 0.170 1.000000e+00 2
## MGRN1 8.334747e-04 1.3557208 0.113 0.185 1.000000e+00 2
## MAPK6.1 8.358113e-04 1.3886284 0.072 0.125 1.000000e+00 2
## IPO11.1 8.382806e-04 1.4276018 0.035 0.071 1.000000e+00 2
## AL080276.2 8.401985e-04 0.1396802 0.008 0.030 1.000000e+00 2
## ZKSCAN5 8.408805e-04 0.6733912 0.023 0.054 1.000000e+00 2
## SLC35E1.1 8.471457e-04 1.2014125 0.075 0.130 1.000000e+00 2
## TMEM143 8.509394e-04 0.4384730 0.020 0.049 1.000000e+00 2
## HOXB-AS1 8.521856e-04 0.4939096 0.009 0.033 1.000000e+00 2
## SIL1.1 8.543387e-04 1.2133143 0.090 0.151 1.000000e+00 2
## ZNF383 8.610137e-04 1.1164904 0.021 0.051 1.000000e+00 2
## PDLIM5.1 8.618750e-04 1.4312609 0.083 0.140 1.000000e+00 2
## PRKAB2 8.630298e-04 0.6993320 0.021 0.051 1.000000e+00 2
## URB2 8.671350e-04 0.7052250 0.012 0.038 1.000000e+00 2
## DMWD 8.683207e-04 0.4558917 0.012 0.038 1.000000e+00 2
## PRKX 8.715661e-04 1.2649474 0.183 0.296 1.000000e+00 2
## UPF2.1 8.742416e-04 1.3328541 0.140 0.225 1.000000e+00 2
## CCR5.2 8.776875e-04 0.7857058 0.036 0.073 1.000000e+00 2
## CLIP3 8.845644e-04 -2.2241506 0.002 0.019 1.000000e+00 2
## SLC49A4 8.850566e-04 0.9139895 0.021 0.051 1.000000e+00 2
## AC023509.6 8.860848e-04 -0.5502169 0.002 0.019 1.000000e+00 2
## AC022182.2.1 8.860848e-04 -1.6400047 0.002 0.019 1.000000e+00 2
## MICU1.1 8.912157e-04 1.3106586 0.086 0.145 1.000000e+00 2
## MTPAP.1 8.928882e-04 1.3803004 0.011 0.035 1.000000e+00 2
## CDCP1 8.975061e-04 0.9928480 0.008 0.030 1.000000e+00 2
## CACTIN 8.997359e-04 1.4488624 0.023 0.054 1.000000e+00 2
## ZNF189 9.082051e-04 0.4685611 0.018 0.047 1.000000e+00 2
## AL451165.2 9.096984e-04 0.5880721 0.009 0.033 1.000000e+00 2
## RECK.1 9.156638e-04 1.2450022 0.035 0.071 1.000000e+00 2
## PRMT9.1 9.169071e-04 1.0824266 0.062 0.109 1.000000e+00 2
## HELQ 9.190146e-04 1.0562790 0.063 0.111 1.000000e+00 2
## RGL2.1 9.202147e-04 1.2571821 0.032 0.067 1.000000e+00 2
## NT5DC2.1 9.239870e-04 0.9599761 0.012 0.037 1.000000e+00 2
## NUDCD3.1 9.257711e-04 1.1391639 0.099 0.164 1.000000e+00 2
## RDH13 9.312443e-04 0.9524907 0.027 0.060 1.000000e+00 2
## ZNF493 9.346064e-04 0.7537793 0.053 0.096 1.000000e+00 2
## NDUFAF5 9.392742e-04 0.7954948 0.033 0.069 1.000000e+00 2
## AC108488.1 9.458643e-04 -0.4480016 0.003 0.022 1.000000e+00 2
## SNURF 9.475460e-04 -0.6305577 0.003 0.022 1.000000e+00 2
## ENOX2 9.489368e-04 1.1983833 0.060 0.107 1.000000e+00 2
## MARS2 9.493116e-04 0.5882455 0.014 0.040 1.000000e+00 2
## TRAV18 9.498275e-04 -1.4325412 0.002 0.019 1.000000e+00 2
## SASH1 9.502423e-04 4.6128116 0.012 0.003 1.000000e+00 2
## AC069224.1 9.509700e-04 4.2925534 0.012 0.003 1.000000e+00 2
## BRAP 9.527751e-04 1.1287221 0.057 0.103 1.000000e+00 2
## ZBTB44 9.560823e-04 1.1385341 0.102 0.168 1.000000e+00 2
## ZNF136 9.624774e-04 0.7324041 0.024 0.055 1.000000e+00 2
## VARS2.1 9.643410e-04 1.2788126 0.017 0.044 1.000000e+00 2
## PIGG 9.692570e-04 0.9393826 0.062 0.109 1.000000e+00 2
## ACAP3 9.710008e-04 1.0605181 0.035 0.071 1.000000e+00 2
## RNF216.1 9.713255e-04 1.2573073 0.149 0.239 1.000000e+00 2
## WDR81 9.762995e-04 1.1791851 0.044 0.084 1.000000e+00 2
## AC131571.1 9.779409e-04 0.8503674 0.018 0.047 1.000000e+00 2
## ANKUB1 9.793982e-04 0.9461093 0.024 0.055 1.000000e+00 2
## PTBP2.1 9.808074e-04 1.1342673 0.089 0.148 1.000000e+00 2
## MTRF1 9.822820e-04 1.1319332 0.039 0.077 1.000000e+00 2
## ARHGAP12 9.832165e-04 1.2548027 0.044 0.084 1.000000e+00 2
## ARFGAP1 9.834414e-04 1.4946107 0.044 0.084 1.000000e+00 2
## RRBP1 9.867611e-04 1.3901085 0.167 0.268 1.000000e+00 2
## TRBV20-1 9.915666e-04 -1.4843847 0.036 0.069 1.000000e+00 2
## ABI1.1 9.923847e-04 1.0562732 0.238 0.389 1.000000e+00 2
## DOK6.1 9.999915e-04 0.7556931 0.026 0.058 1.000000e+00 2
## TRAV21 1.001938e-03 -1.2400082 0.023 0.052 1.000000e+00 2
## PLCXD1 1.002894e-03 0.7344705 0.014 0.039 1.000000e+00 2
## AC008063.1.1 1.003954e-03 -1.2289412 0.005 0.025 1.000000e+00 2
## MIER3 1.005335e-03 1.3277238 0.023 0.053 1.000000e+00 2
## LPAR5.1 1.005640e-03 0.7260995 0.024 0.055 1.000000e+00 2
## MAP4K3-DT 1.009485e-03 0.6740978 0.011 0.035 1.000000e+00 2
## CDKL3.1 1.010243e-03 0.8512085 0.018 0.046 1.000000e+00 2
## ZNF486 1.011292e-03 0.1993442 0.006 0.027 1.000000e+00 2
## TEX10 1.014546e-03 1.3126833 0.054 0.099 1.000000e+00 2
## PELI3 1.015849e-03 -2.2988550 0.002 0.019 1.000000e+00 2
## LY6E-DT 1.016237e-03 -2.4504811 0.002 0.019 1.000000e+00 2
## CHML 1.016633e-03 -1.2564470 0.005 0.025 1.000000e+00 2
## ZNF485 1.016879e-03 -3.9233529 0.000 0.016 1.000000e+00 2
## ARL11 1.016879e-03 -4.0008946 0.000 0.016 1.000000e+00 2
## TLCD3A 1.018176e-03 -1.7108765 0.002 0.019 1.000000e+00 2
## CCDC77.1 1.019060e-03 1.3213671 0.029 0.062 1.000000e+00 2
## AGO1 1.019653e-03 0.9117175 0.026 0.058 1.000000e+00 2
## AP1G1.1 1.020921e-03 1.4804979 0.122 0.197 1.000000e+00 2
## NDOR1.1 1.022138e-03 1.1274585 0.030 0.064 1.000000e+00 2
## MTHFSD.1 1.031340e-03 0.7305640 0.018 0.046 1.000000e+00 2
## NUS1 1.031817e-03 1.2802266 0.110 0.179 1.000000e+00 2
## ZNF605 1.032092e-03 0.2301901 0.008 0.030 1.000000e+00 2
## MED22 1.034047e-03 0.6501560 0.009 0.032 1.000000e+00 2
## CCNL2 1.037806e-03 1.3237211 0.128 0.207 1.000000e+00 2
## KLHL8.1 1.039592e-03 0.9913313 0.032 0.066 1.000000e+00 2
## DENND1B.1 1.046458e-03 1.8867774 0.305 0.334 1.000000e+00 2
## TRAV26-2.1 1.053441e-03 -0.4174407 0.011 0.034 1.000000e+00 2
## SLF2.1 1.054763e-03 1.2845791 0.090 0.150 1.000000e+00 2
## PPP2R2A.1 1.061820e-03 1.2051289 0.122 0.197 1.000000e+00 2
## ZNF141.1 1.064467e-03 1.2842930 0.056 0.100 1.000000e+00 2
## NUP188.2 1.069221e-03 1.0203690 0.060 0.107 1.000000e+00 2
## TRBV4-2.1 1.069963e-03 -1.9042146 0.020 0.047 1.000000e+00 2
## MIER2.1 1.076306e-03 1.1749056 0.018 0.046 1.000000e+00 2
## CLTC 1.076771e-03 1.1956416 0.191 0.306 1.000000e+00 2
## ZNF845 1.080690e-03 0.4216242 0.011 0.034 1.000000e+00 2
## PEX13 1.084009e-03 1.2557584 0.057 0.103 1.000000e+00 2
## CLCN7 1.085254e-03 1.3278515 0.041 0.079 1.000000e+00 2
## EHD4 1.089937e-03 0.9035397 0.029 0.062 1.000000e+00 2
## C3orf33 1.091093e-03 -3.8742295 0.000 0.016 1.000000e+00 2
## AP003307.1 1.092716e-03 -1.0542816 0.002 0.019 1.000000e+00 2
## GPR150 1.092716e-03 -0.4662023 0.002 0.019 1.000000e+00 2
## ARL17B.1 1.095614e-03 1.0534408 0.035 0.070 1.000000e+00 2
## AC019163.1 1.098061e-03 -0.2196746 0.005 0.024 1.000000e+00 2
## MINCR 1.099173e-03 0.2553083 0.009 0.032 1.000000e+00 2
## TIGD1 1.100661e-03 0.4702369 0.008 0.030 1.000000e+00 2
## EMSY 1.101549e-03 1.2755072 0.099 0.163 1.000000e+00 2
## RAB2B 1.104234e-03 0.9593412 0.039 0.077 1.000000e+00 2
## ZNF142 1.109659e-03 0.6794685 0.027 0.059 1.000000e+00 2
## IL2RA.1 1.110469e-03 1.1665629 0.179 0.283 1.000000e+00 2
## KDM5D.1 1.112135e-03 1.2326646 0.081 0.137 1.000000e+00 2
## KANSL1.1 1.115151e-03 1.7685696 0.332 0.378 1.000000e+00 2
## PAXIP1.2 1.117324e-03 1.1421898 0.029 0.062 1.000000e+00 2
## EXT1.2 1.129599e-03 2.4991397 0.138 0.109 1.000000e+00 2
## WDR3 1.130860e-03 1.4287899 0.086 0.143 1.000000e+00 2
## ATXN7L3 1.131280e-03 1.1749130 0.047 0.087 1.000000e+00 2
## USP12 1.138209e-03 1.1495016 0.084 0.141 1.000000e+00 2
## FAM229B 1.141098e-03 0.5981279 0.014 0.039 1.000000e+00 2
## DHFR2 1.141352e-03 -0.5838221 0.009 0.032 1.000000e+00 2
## MYCBP2-AS1 1.141470e-03 0.6276946 0.017 0.044 1.000000e+00 2
## TSPAN5.1 1.141756e-03 1.2345589 0.192 0.308 1.000000e+00 2
## TMEM161B.1 1.145771e-03 1.1832896 0.071 0.122 1.000000e+00 2
## TANK 1.146996e-03 1.6073513 0.368 0.443 1.000000e+00 2
## ANKRD35.1 1.152813e-03 -1.8644971 0.003 0.022 1.000000e+00 2
## EID2B 1.163882e-03 1.3424430 0.018 0.046 1.000000e+00 2
## GTF2IRD2B 1.165330e-03 0.7315614 0.032 0.066 1.000000e+00 2
## GAPVD1 1.166499e-03 1.3893426 0.143 0.229 1.000000e+00 2
## ETAA1 1.168644e-03 0.7381849 0.032 0.066 1.000000e+00 2
## FADS1 1.170505e-03 -1.3742938 0.002 0.019 1.000000e+00 2
## ZNF57 1.170505e-03 -1.9088929 0.002 0.019 1.000000e+00 2
## ZNRF2 1.172335e-03 1.2441993 0.126 0.203 1.000000e+00 2
## MRPL30 1.172397e-03 -0.6125503 0.005 0.024 1.000000e+00 2
## RUNX3.1 1.182163e-03 1.1708342 0.245 0.399 1.000000e+00 2
## AC104162.2.1 1.182543e-03 0.9680342 0.027 0.059 1.000000e+00 2
## ZNF814 1.188593e-03 0.9807054 0.035 0.070 1.000000e+00 2
## RALGDS.1 1.189983e-03 1.2429526 0.174 0.278 1.000000e+00 2
## NCKIPSD.1 1.193150e-03 0.6749263 0.023 0.052 1.000000e+00 2
## TMEM234 1.197623e-03 0.5826560 0.015 0.041 1.000000e+00 2
## ARMC8 1.202256e-03 1.1786603 0.119 0.192 1.000000e+00 2
## ACO1 1.204661e-03 0.5878012 0.014 0.039 1.000000e+00 2
## MED29 1.209362e-03 1.6449764 0.119 0.192 1.000000e+00 2
## LONP2 1.214114e-03 1.1527626 0.208 0.332 1.000000e+00 2
## SGPP1.1 1.214499e-03 1.2424728 0.059 0.104 1.000000e+00 2
## APBB3 1.216106e-03 0.7072649 0.017 0.043 1.000000e+00 2
## SMARCAD1.1 1.219825e-03 1.2564416 0.099 0.162 1.000000e+00 2
## GCA.2 1.220956e-03 1.1691775 0.027 0.059 1.000000e+00 2
## MICA 1.224121e-03 0.5054093 0.011 0.034 1.000000e+00 2
## PIP5K1C 1.232018e-03 0.7655086 0.023 0.052 1.000000e+00 2
## LINC02256.1 1.233029e-03 0.2589471 0.006 0.027 1.000000e+00 2
## ATP8A1.1 1.233727e-03 1.9171219 0.229 0.218 1.000000e+00 2
## TRAV20.1 1.235416e-03 -1.8869672 0.006 0.026 1.000000e+00 2
## BRF1.2 1.235695e-03 1.1973070 0.057 0.102 1.000000e+00 2
## HECW2-AS1.1 1.240960e-03 0.9947001 0.030 0.063 1.000000e+00 2
## AP001816.1 1.242555e-03 0.6139321 0.009 0.032 1.000000e+00 2
## STX12 1.243604e-03 1.2993162 0.101 0.165 1.000000e+00 2
## KLRC1.2 1.246695e-03 -0.4361615 0.011 0.034 1.000000e+00 2
## ASB13.1 1.249091e-03 -2.5285628 0.002 0.018 1.000000e+00 2
## LMF1 1.249866e-03 1.2686906 0.068 0.117 1.000000e+00 2
## FAM83D.2 1.252200e-03 0.3771851 0.017 0.043 1.000000e+00 2
## CCDC65.1 1.253888e-03 0.3937357 0.008 0.029 1.000000e+00 2
## RBKS 1.255327e-03 0.4860798 0.012 0.036 1.000000e+00 2
## AL162258.2 1.256730e-03 -0.6047985 0.002 0.018 1.000000e+00 2
## KDM5A.1 1.261794e-03 1.0831819 0.223 0.358 1.000000e+00 2
## TAF13 1.263649e-03 1.2988033 0.032 0.066 1.000000e+00 2
## MED23 1.266856e-03 1.2387526 0.099 0.163 1.000000e+00 2
## LIN54.2 1.268744e-03 1.4598135 0.081 0.136 1.000000e+00 2
## TSPAN18.1 1.273108e-03 0.9350015 0.039 0.076 1.000000e+00 2
## ZCCHC2.1 1.278073e-03 1.2434636 0.086 0.143 1.000000e+00 2
## ZFP1 1.282048e-03 0.6716052 0.014 0.039 1.000000e+00 2
## SLAMF1.1 1.282600e-03 1.0611059 0.227 0.364 1.000000e+00 2
## F8 1.289097e-03 0.7389843 0.014 0.039 1.000000e+00 2
## POLR3D 1.303786e-03 1.1296988 0.044 0.082 1.000000e+00 2
## CCSAP.1 1.305130e-03 1.1367444 0.026 0.057 1.000000e+00 2
## TRRAP.1 1.309439e-03 1.2489211 0.126 0.202 1.000000e+00 2
## F2R.1 1.313216e-03 0.8605261 0.044 0.082 1.000000e+00 2
## SLC39A14.1 1.323472e-03 0.9544034 0.021 0.050 1.000000e+00 2
## MPZL1.1 1.327537e-03 0.5191539 0.009 0.031 1.000000e+00 2
## TMEM245 1.328888e-03 1.0368007 0.090 0.149 1.000000e+00 2
## TSEN2 1.329087e-03 1.1874982 0.023 0.052 1.000000e+00 2
## ZNF93.2 1.330604e-03 0.4058429 0.008 0.029 1.000000e+00 2
## MIA2.2 1.332082e-03 1.2607037 0.185 0.294 1.000000e+00 2
## ZC3H7B 1.332677e-03 1.1650097 0.044 0.082 1.000000e+00 2
## REXO5 1.332987e-03 -3.1070757 0.002 0.018 1.000000e+00 2
## JARID2.1 1.333035e-03 1.9761371 0.286 0.304 1.000000e+00 2
## BDP1.1 1.334942e-03 1.1062990 0.245 0.396 1.000000e+00 2
## C1orf56.1 1.335108e-03 -1.0469138 0.003 0.021 1.000000e+00 2
## ADAL.1 1.343067e-03 1.3876880 0.020 0.048 1.000000e+00 2
## TCFL5 1.343555e-03 0.4375970 0.023 0.052 1.000000e+00 2
## TMEM156.1 1.344165e-03 1.3144346 0.114 0.184 1.000000e+00 2
## AC105446.1 1.345834e-03 -0.3553654 0.005 0.024 1.000000e+00 2
## EXTL2 1.347177e-03 -0.7774204 0.005 0.024 1.000000e+00 2
## AL161421.1 1.347797e-03 -0.3747791 0.002 0.018 1.000000e+00 2
## AL109628.2 1.348521e-03 -0.7783189 0.005 0.024 1.000000e+00 2
## AACS.1 1.359931e-03 0.8943745 0.024 0.054 1.000000e+00 2
## SLC25A37.2 1.361741e-03 1.5419564 0.035 0.070 1.000000e+00 2
## CEPT1.1 1.362638e-03 1.0749823 0.080 0.134 1.000000e+00 2
## MARCH7 1.365746e-03 1.1855989 0.189 0.302 1.000000e+00 2
## ABCD2.1 1.370051e-03 1.1762035 0.165 0.261 1.000000e+00 2
## HS2ST1.1 1.374426e-03 1.4523651 0.093 0.153 1.000000e+00 2
## TTC27.1 1.383926e-03 1.2103088 0.060 0.106 1.000000e+00 2
## ARHGAP21.1 1.391591e-03 0.8601444 0.029 0.061 1.000000e+00 2
## EOMES.1 1.392846e-03 0.4968630 0.038 0.073 1.000000e+00 2
## SETBP1.2 1.400899e-03 2.7447686 0.045 0.025 1.000000e+00 2
## EIF3J-DT 1.402915e-03 1.4935494 0.032 0.065 1.000000e+00 2
## GTF2H2 1.406455e-03 0.4071411 0.006 0.026 1.000000e+00 2
## KIF20A.2 1.409464e-03 -0.4257353 0.005 0.024 1.000000e+00 2
## MPP5.1 1.415295e-03 1.3729533 0.033 0.067 1.000000e+00 2
## CRK.1 1.419553e-03 1.2679605 0.042 0.080 1.000000e+00 2
## ATP10A.1 1.421375e-03 1.9722296 0.250 0.247 1.000000e+00 2
## ATG10.1 1.421938e-03 1.1695877 0.080 0.133 1.000000e+00 2
## AKAP11 1.425263e-03 1.1224739 0.108 0.175 1.000000e+00 2
## TSTD2 1.428813e-03 1.1869116 0.044 0.082 1.000000e+00 2
## CKB 1.431717e-03 -0.6987518 0.003 0.021 1.000000e+00 2
## VPS39 1.432881e-03 1.3875316 0.086 0.142 1.000000e+00 2
## TMEM104 1.433934e-03 1.0485447 0.036 0.071 1.000000e+00 2
## DROSHA.1 1.434674e-03 1.2031063 0.063 0.110 1.000000e+00 2
## KIAA0930 1.435373e-03 0.4503881 0.014 0.038 1.000000e+00 2
## HBEGF 1.439457e-03 -2.4115411 0.002 0.018 1.000000e+00 2
## SBF2-AS1.1 1.441669e-03 -0.5479802 0.005 0.024 1.000000e+00 2
## SLC22A5.1 1.442627e-03 -0.1032947 0.005 0.024 1.000000e+00 2
## PWWP2B 1.443106e-03 -0.4732234 0.005 0.024 1.000000e+00 2
## TMEM191C 1.446714e-03 -3.9695448 0.000 0.015 1.000000e+00 2
## ENTPD3-AS1.1 1.447552e-03 0.8418788 0.014 0.038 1.000000e+00 2
## XRN1.1 1.449234e-03 0.9997123 0.289 0.478 1.000000e+00 2
## ZNF526 1.450661e-03 0.9249917 0.024 0.054 1.000000e+00 2
## CYTH4.1 1.453067e-03 1.2273158 0.140 0.222 1.000000e+00 2
## FAM49B.1 1.456671e-03 1.0571556 0.284 0.473 1.000000e+00 2
## GXYLT1.2 1.461291e-03 0.8746063 0.039 0.076 1.000000e+00 2
## ZNF268 1.463957e-03 0.7153747 0.020 0.047 1.000000e+00 2
## TRMT2B.1 1.472953e-03 1.0632523 0.032 0.065 1.000000e+00 2
## CASC1 1.473652e-03 0.4817124 0.011 0.034 1.000000e+00 2
## CLPB.1 1.474214e-03 1.3851095 0.036 0.071 1.000000e+00 2
## CDC73.1 1.475536e-03 1.2983655 0.221 0.357 1.000000e+00 2
## PLCG1 1.478112e-03 1.2512523 0.211 0.338 1.000000e+00 2
## CDK5RAP2.2 1.479986e-03 1.1142604 0.111 0.178 1.000000e+00 2
## CSKMT.1 1.481240e-03 2.7842290 0.102 0.074 1.000000e+00 2
## BTBD10 1.484037e-03 1.2554599 0.116 0.186 1.000000e+00 2
## TMEM161B-AS1 1.488395e-03 1.1003803 0.060 0.106 1.000000e+00 2
## N4BP2.1 1.492723e-03 1.3875138 0.156 0.248 1.000000e+00 2
## RELT.1 1.492912e-03 1.3295888 0.056 0.099 1.000000e+00 2
## RB1CC1.1 1.493252e-03 1.2063412 0.176 0.280 1.000000e+00 2
## ARL6 1.494038e-03 0.1748719 0.006 0.026 1.000000e+00 2
## AC096677.1 1.497898e-03 0.6171889 0.017 0.043 1.000000e+00 2
## AC253572.2.1 1.499350e-03 1.0634757 0.123 0.196 1.000000e+00 2
## TMEM67 1.503753e-03 0.3920411 0.009 0.031 1.000000e+00 2
## ZEB1-AS1.1 1.507220e-03 0.2788903 0.009 0.031 1.000000e+00 2
## AC008443.1 1.512412e-03 0.6190329 0.021 0.049 1.000000e+00 2
## TBCEL 1.519182e-03 0.8133786 0.029 0.060 1.000000e+00 2
## SNX19 1.519556e-03 1.1298475 0.065 0.112 1.000000e+00 2
## FPGT.1 1.523789e-03 1.1790338 0.020 0.047 1.000000e+00 2
## ADCY3.1 1.527849e-03 1.0966338 0.023 0.052 1.000000e+00 2
## POLE.2 1.530162e-03 1.3108080 0.042 0.080 1.000000e+00 2
## ANKEF1 1.530616e-03 0.2437381 0.014 0.038 1.000000e+00 2
## ZNF654.1 1.531899e-03 1.1970258 0.093 0.153 1.000000e+00 2
## ZNF280C 1.540750e-03 0.8398086 0.017 0.043 1.000000e+00 2
## HEXIM2 1.541287e-03 0.8434515 0.011 0.033 1.000000e+00 2
## DGCR8.1 1.544131e-03 1.0020902 0.036 0.071 1.000000e+00 2
## AC002310.1 1.545325e-03 -0.1664929 0.005 0.023 1.000000e+00 2
## S100A5 1.552578e-03 -4.0120601 0.000 0.015 1.000000e+00 2
## IQGAP3.2 1.552578e-03 -4.2585106 0.000 0.015 1.000000e+00 2
## PRKAG2-AS1.1 1.552578e-03 -3.8904593 0.000 0.015 1.000000e+00 2
## TUBGCP5 1.556011e-03 1.2467250 0.045 0.084 1.000000e+00 2
## KDM3A 1.559114e-03 1.1553656 0.098 0.159 1.000000e+00 2
## DNAJC27 1.560712e-03 1.0349582 0.032 0.065 1.000000e+00 2
## DHX37 1.564982e-03 0.5107465 0.020 0.047 1.000000e+00 2
## UPF1.1 1.569196e-03 1.4676648 0.084 0.140 1.000000e+00 2
## PRKD2 1.570009e-03 1.2225878 0.141 0.223 1.000000e+00 2
## LINC01176 1.575445e-03 4.2389073 0.014 0.004 1.000000e+00 2
## TPRG1L 1.576260e-03 0.7372417 0.033 0.067 1.000000e+00 2
## YPEL1.1 1.588740e-03 0.7493828 0.021 0.049 1.000000e+00 2
## POLK.1 1.591810e-03 1.2754120 0.104 0.168 1.000000e+00 2
## PI4K2B.1 1.591996e-03 1.1971453 0.084 0.139 1.000000e+00 2
## IGFBP4.2 1.594366e-03 0.4955300 0.009 0.031 1.000000e+00 2
## TRBV6-5.1 1.597339e-03 -1.7581477 0.009 0.030 1.000000e+00 2
## FAM241A.2 1.597580e-03 0.9830179 0.020 0.047 1.000000e+00 2
## MYB.2 1.604087e-03 0.2436265 0.006 0.026 1.000000e+00 2
## SHLD3 1.606102e-03 0.6297726 0.006 0.026 1.000000e+00 2
## CRTAM.2 1.606102e-03 0.5045185 0.006 0.026 1.000000e+00 2
## SH3BP5-AS1 1.612765e-03 0.3405619 0.012 0.036 1.000000e+00 2
## ZNF777 1.613935e-03 1.0177056 0.023 0.051 1.000000e+00 2
## HMGCR 1.614073e-03 1.1853322 0.063 0.109 1.000000e+00 2
## AC008083.3.1 1.622870e-03 0.8770806 0.030 0.062 1.000000e+00 2
## SLC25A4 1.624515e-03 0.5317998 0.008 0.028 1.000000e+00 2
## HIST1H2BD.2 1.635405e-03 2.7465406 0.080 0.054 1.000000e+00 2
## DDIT4-AS1.1 1.637741e-03 -1.0061764 0.003 0.021 1.000000e+00 2
## ZNF746 1.640693e-03 1.4489711 0.051 0.092 1.000000e+00 2
## AC007383.2 1.640814e-03 0.2374172 0.011 0.033 1.000000e+00 2
## SERGEF 1.641512e-03 1.2100589 0.069 0.118 1.000000e+00 2
## AC097382.3 1.641787e-03 -1.1350522 0.003 0.021 1.000000e+00 2
## CRIP2.1 1.642945e-03 -0.3763353 0.003 0.021 1.000000e+00 2
## SBK1.1 1.643293e-03 -1.3039366 0.005 0.023 1.000000e+00 2
## KLHL24.1 1.644456e-03 1.1001274 0.081 0.135 1.000000e+00 2
## ZNF529 1.651427e-03 1.0262398 0.042 0.079 1.000000e+00 2
## MTMR9 1.659974e-03 0.7542098 0.021 0.049 1.000000e+00 2
## IL18R1.1 1.660708e-03 1.1615994 0.069 0.117 1.000000e+00 2
## CLASP2 1.661928e-03 1.3809963 0.138 0.220 1.000000e+00 2
## AC009950.1 1.662789e-03 -1.3320924 0.002 0.018 1.000000e+00 2
## SLC35F1 1.662789e-03 -1.0912600 0.002 0.018 1.000000e+00 2
## MEGF9 1.666254e-03 1.0718936 0.084 0.138 1.000000e+00 2
## MRPL53 1.666256e-03 -3.9091487 0.000 0.015 1.000000e+00 2
## UBE3D 1.671681e-03 1.1376960 0.032 0.064 1.000000e+00 2
## QSER1.2 1.674012e-03 1.0887872 0.032 0.064 1.000000e+00 2
## VPS18.1 1.675776e-03 1.3548757 0.027 0.058 1.000000e+00 2
## HYLS1 1.676369e-03 1.2759355 0.026 0.056 1.000000e+00 2
## ZNF83.2 1.676658e-03 1.1319818 0.090 0.147 1.000000e+00 2
## PDE4B.1 1.678117e-03 1.1645975 0.152 0.239 1.000000e+00 2
## SEMA4D 1.680502e-03 1.5529609 0.377 0.466 1.000000e+00 2
## ZMIZ2.1 1.690487e-03 1.3779627 0.056 0.099 1.000000e+00 2
## LIX1L-AS1 1.692733e-03 1.0747756 0.026 0.056 1.000000e+00 2
## KLF7.1 1.698202e-03 0.9963597 0.027 0.058 1.000000e+00 2
## ZMAT1 1.700898e-03 0.5094426 0.014 0.038 1.000000e+00 2
## VDR.1 1.706316e-03 0.4728507 0.012 0.035 1.000000e+00 2
## DNMT3A 1.711256e-03 1.1872155 0.092 0.150 1.000000e+00 2
## USP46.1 1.726943e-03 1.3226371 0.068 0.116 1.000000e+00 2
## UBFD1.1 1.727298e-03 1.0412789 0.035 0.069 1.000000e+00 2
## NUMB.1 1.739650e-03 1.0868093 0.117 0.186 1.000000e+00 2
## NOL6 1.743759e-03 0.8226398 0.015 0.040 1.000000e+00 2
## CD84.1 1.748014e-03 1.1887610 0.084 0.138 1.000000e+00 2
## SETD9 1.751239e-03 0.7503854 0.011 0.033 1.000000e+00 2
## EPM2A.1 1.751947e-03 -1.0293067 0.003 0.020 1.000000e+00 2
## MCM3AP 1.752472e-03 1.4415706 0.099 0.160 1.000000e+00 2
## KLHL28 1.752498e-03 1.1941737 0.092 0.150 1.000000e+00 2
## TLCD1.2 1.756075e-03 0.6166792 0.011 0.033 1.000000e+00 2
## FBXO43.2 1.757862e-03 -3.2516493 0.002 0.018 1.000000e+00 2
## MANBA 1.769875e-03 1.2394621 0.084 0.139 1.000000e+00 2
## XXYLT1.1 1.776026e-03 1.0867469 0.033 0.066 1.000000e+00 2
## GNB4 1.788331e-03 -3.9603250 0.000 0.014 1.000000e+00 2
## NREP.2 1.788331e-03 -3.7549040 0.000 0.014 1.000000e+00 2
## AC123768.5 1.788331e-03 -3.8754281 0.000 0.014 1.000000e+00 2
## PPP1R16B.1 1.812544e-03 2.0296236 0.230 0.224 1.000000e+00 2
## LINC00920 1.820016e-03 0.8714366 0.009 0.030 1.000000e+00 2
## MTFR1.1 1.821123e-03 1.3432450 0.077 0.128 1.000000e+00 2
## PPP1R16A 1.822480e-03 0.8205941 0.014 0.038 1.000000e+00 2
## ZNF460 1.824456e-03 1.0991875 0.063 0.108 1.000000e+00 2
## AC073254.1 1.826414e-03 -0.4716241 0.006 0.026 1.000000e+00 2
## NAB1.1 1.831120e-03 0.7835167 0.017 0.042 1.000000e+00 2
## KLHL18.2 1.835198e-03 1.2833356 0.062 0.107 1.000000e+00 2
## KIAA0586.1 1.838688e-03 1.2383960 0.105 0.169 1.000000e+00 2
## SLC66A1.1 1.846539e-03 1.2756109 0.017 0.042 1.000000e+00 2
## RANBP10 1.846796e-03 1.4822899 0.057 0.100 1.000000e+00 2
## ZNF420 1.868510e-03 0.9572884 0.033 0.066 1.000000e+00 2
## TMEM94 1.873161e-03 1.1796252 0.041 0.077 1.000000e+00 2
## ANAPC2 1.877237e-03 1.3831206 0.032 0.064 1.000000e+00 2
## POGZ.2 1.879769e-03 1.4071833 0.083 0.136 1.000000e+00 2
## MAMSTR 1.882726e-03 -0.8753849 0.003 0.020 1.000000e+00 2
## ICAM1 1.883543e-03 1.4751687 0.066 0.113 1.000000e+00 2
## AC009630.1 1.884833e-03 -0.3940022 0.005 0.023 1.000000e+00 2
## CBX8 1.893777e-03 0.3994478 0.011 0.033 1.000000e+00 2
## TRBV24-1 1.895431e-03 -2.6590406 0.005 0.023 1.000000e+00 2
## TYMSOS.1 1.912226e-03 -1.3982772 0.002 0.017 1.000000e+00 2
## LPAR6.1 1.912392e-03 1.2463808 0.075 0.125 1.000000e+00 2
## HMGN5 1.912953e-03 -0.1161214 0.002 0.017 1.000000e+00 2
## FAM135A 1.914677e-03 0.6665907 0.020 0.046 1.000000e+00 2
## TMSB4Y 1.919433e-03 -4.0737236 0.000 0.014 1.000000e+00 2
## SLC9A3R2 1.919433e-03 -3.9987800 0.000 0.014 1.000000e+00 2
## WDR41 1.929799e-03 1.0021333 0.045 0.083 1.000000e+00 2
## HIST1H3C.2 1.934792e-03 0.9735833 0.024 0.053 1.000000e+00 2
## TMEM19 1.935211e-03 0.7274775 0.024 0.053 1.000000e+00 2
## CASP9 1.939477e-03 1.2784869 0.036 0.070 1.000000e+00 2
## C1orf21.1 1.942687e-03 0.5753886 0.009 0.030 1.000000e+00 2
## UBAP1.1 1.944439e-03 1.2102801 0.128 0.202 1.000000e+00 2
## EIF3C 1.944904e-03 0.6645091 0.009 0.030 1.000000e+00 2
## MAP1LC3A 1.953978e-03 0.2697879 0.006 0.025 1.000000e+00 2
## MLLT1.1 1.954119e-03 0.9628492 0.024 0.053 1.000000e+00 2
## ADAP1.1 1.954369e-03 1.0653924 0.021 0.049 1.000000e+00 2
## SLC30A4 1.958252e-03 -0.1792816 0.006 0.025 1.000000e+00 2
## GLMN.1 1.958512e-03 1.3770628 0.110 0.176 1.000000e+00 2
## LRRC20.1 1.967744e-03 0.5960502 0.008 0.028 1.000000e+00 2
## PROSER3 1.971633e-03 1.0548557 0.020 0.046 1.000000e+00 2
## PIP5K1A.1 1.972794e-03 1.2500225 0.069 0.117 1.000000e+00 2
## TMCO3 1.974688e-03 1.2889657 0.048 0.087 1.000000e+00 2
## PIAS2.1 1.977060e-03 1.1053503 0.051 0.091 1.000000e+00 2
## SLC1A4.1 1.982314e-03 1.3535113 0.068 0.114 1.000000e+00 2
## ARMC5 1.984302e-03 1.5514842 0.029 0.059 1.000000e+00 2
## MKS1 1.989560e-03 0.7814265 0.018 0.044 1.000000e+00 2
## OPA3.1 1.993270e-03 1.3598550 0.056 0.098 1.000000e+00 2
## KLF5.2 1.993319e-03 0.5455763 0.011 0.033 1.000000e+00 2
## LRRC28.1 1.995604e-03 1.4533218 0.048 0.087 1.000000e+00 2
## LRATD2 1.997119e-03 -1.6724271 0.003 0.020 1.000000e+00 2
## HABP4.1 1.997715e-03 1.3483429 0.032 0.064 1.000000e+00 2
## ZBTB1 1.998661e-03 1.3124063 0.183 0.290 1.000000e+00 2
## YAF2 2.001223e-03 1.2763100 0.107 0.171 1.000000e+00 2
## PTPN9.1 2.014987e-03 1.3408854 0.110 0.175 1.000000e+00 2
## AC135457.1 2.022879e-03 -0.5524149 0.005 0.023 1.000000e+00 2
## DAAM1 2.023160e-03 1.2415319 0.059 0.102 1.000000e+00 2
## PIK3CG.1 2.036530e-03 1.1578288 0.113 0.179 1.000000e+00 2
## C8orf58 2.060238e-03 -3.9576086 0.000 0.014 1.000000e+00 2
## ANG.1 2.060238e-03 -3.8006949 0.000 0.014 1.000000e+00 2
## C19orf57.2 2.060238e-03 -3.5225240 0.000 0.014 1.000000e+00 2
## ZNF680 2.070207e-03 1.1916449 0.027 0.057 1.000000e+00 2
## GOLGA3 2.076836e-03 1.3112781 0.099 0.160 1.000000e+00 2
## DICER1 2.109131e-03 1.3449789 0.117 0.186 1.000000e+00 2
## TIPARP 2.109593e-03 1.2010805 0.158 0.248 1.000000e+00 2
## NCOA6 2.117520e-03 1.2782147 0.111 0.178 1.000000e+00 2
## PRH1.1 2.129734e-03 1.2849581 0.066 0.112 1.000000e+00 2
## ZNF687.1 2.136952e-03 0.3638661 0.012 0.035 1.000000e+00 2
## STK11IP 2.140978e-03 1.0470262 0.011 0.032 1.000000e+00 2
## LRFN3 2.142735e-03 0.1930331 0.011 0.032 1.000000e+00 2
## TMEM229B 2.146460e-03 1.2059601 0.021 0.048 1.000000e+00 2
## TMSB15B.2 2.155667e-03 0.2840662 0.012 0.035 1.000000e+00 2
## C17orf67 2.156506e-03 0.9593427 0.027 0.057 1.000000e+00 2
## NANP 2.161006e-03 0.1535595 0.005 0.022 1.000000e+00 2
## OR4D1 2.161717e-03 -0.1156821 0.005 0.022 1.000000e+00 2
## C17orf80 2.166173e-03 0.8250670 0.024 0.053 1.000000e+00 2
## PYROXD2 2.166558e-03 0.8937828 0.027 0.057 1.000000e+00 2
## CARF 2.168719e-03 0.9660564 0.021 0.048 1.000000e+00 2
## KIAA1109 2.169932e-03 1.9540153 0.272 0.285 1.000000e+00 2
## RHEBL1.1 2.171184e-03 1.0117159 0.017 0.042 1.000000e+00 2
## CCDC6.1 2.173519e-03 1.3973630 0.078 0.129 1.000000e+00 2
## NBPF15 2.177306e-03 0.6630939 0.027 0.057 1.000000e+00 2
## RBM26 2.184484e-03 1.1772418 0.205 0.323 1.000000e+00 2
## DNHD1 2.196594e-03 1.4572726 0.021 0.048 1.000000e+00 2
## ZNF839 2.198893e-03 1.1432436 0.029 0.059 1.000000e+00 2
## AC145207.2.1 2.200175e-03 -1.6134493 0.002 0.017 1.000000e+00 2
## IGFBP6 2.201011e-03 -0.9661998 0.002 0.017 1.000000e+00 2
## CHRM3-AS2 2.201570e-03 1.0296979 0.054 0.095 1.000000e+00 2
## RCL1 2.206560e-03 1.1106197 0.054 0.095 1.000000e+00 2
## SUFU.1 2.210008e-03 1.2244253 0.050 0.089 1.000000e+00 2
## HOOK1.2 2.211474e-03 -3.8233208 0.000 0.014 1.000000e+00 2
## ERI1.1 2.216034e-03 1.3397293 0.120 0.189 1.000000e+00 2
## DIXDC1.2 2.224410e-03 0.2097410 0.006 0.025 1.000000e+00 2
## PLAUR.1 2.231339e-03 0.5261353 0.006 0.025 1.000000e+00 2
## TGFBR3.1 2.232291e-03 2.4710040 0.116 0.089 1.000000e+00 2
## NAA35 2.236074e-03 1.2065114 0.104 0.166 1.000000e+00 2
## GTF3C1 2.236568e-03 1.2333963 0.104 0.166 1.000000e+00 2
## FAM20B.1 2.239259e-03 1.4251773 0.038 0.072 1.000000e+00 2
## AC018816.1 2.239527e-03 1.5179007 0.099 0.159 1.000000e+00 2
## DCAF6 2.244112e-03 1.1096914 0.123 0.195 1.000000e+00 2
## YLPM1.1 2.257533e-03 1.2095772 0.156 0.245 1.000000e+00 2
## EOGT.1 2.257952e-03 0.4616634 0.012 0.034 1.000000e+00 2
## AC002467.1.1 2.258881e-03 0.4270887 0.011 0.032 1.000000e+00 2
## OTUD4 2.266050e-03 1.2867162 0.102 0.164 1.000000e+00 2
## FLVCR1-DT 2.272416e-03 1.0449782 0.018 0.044 1.000000e+00 2
## FLYWCH1 2.276120e-03 1.1774555 0.045 0.082 1.000000e+00 2
## C1orf54 2.277217e-03 0.6937585 0.018 0.044 1.000000e+00 2
## AJM1 2.286031e-03 1.5526398 0.023 0.050 1.000000e+00 2
## PPP1R37 2.292213e-03 1.0625495 0.018 0.044 1.000000e+00 2
## RNF38.1 2.292607e-03 1.9155964 0.256 0.261 1.000000e+00 2
## TBCCD1.1 2.297770e-03 1.4212419 0.060 0.103 1.000000e+00 2
## ZNF251 2.298190e-03 0.7265222 0.033 0.065 1.000000e+00 2
## FSD1 2.314614e-03 0.6455622 0.005 0.022 1.000000e+00 2
## MBLAC2 2.326274e-03 0.4234489 0.014 0.037 1.000000e+00 2
## IKZF3.2 2.331600e-03 1.1645342 0.189 0.297 1.000000e+00 2
## AC073529.1.2 2.353797e-03 -0.1376717 0.006 0.025 1.000000e+00 2
## EFCAB7 2.354580e-03 1.2351634 0.026 0.055 1.000000e+00 2
## PVRIG.1 2.359237e-03 -1.6754218 0.002 0.017 1.000000e+00 2
## PGM3.1 2.361796e-03 0.8141352 0.023 0.050 1.000000e+00 2
## ABHD18 2.363626e-03 1.4068055 0.117 0.186 1.000000e+00 2
## ALS2 2.372324e-03 0.9981399 0.033 0.065 1.000000e+00 2
## EPHX2.1 2.381002e-03 0.4173059 0.006 0.025 1.000000e+00 2
## FAM171A1.2 2.385440e-03 0.5245404 0.006 0.025 1.000000e+00 2
## ALDH5A1 2.394581e-03 0.3546548 0.008 0.027 1.000000e+00 2
## RAB11FIP4 2.405131e-03 0.8970542 0.051 0.090 1.000000e+00 2
## MSANTD2 2.416460e-03 0.5617420 0.023 0.050 1.000000e+00 2
## STRBP.2 2.420816e-03 1.2950695 0.054 0.095 1.000000e+00 2
## KIAA1841.1 2.425708e-03 0.9703632 0.023 0.050 1.000000e+00 2
## PPP1R9B 2.428831e-03 1.2885823 0.056 0.097 1.000000e+00 2
## KIF16B.2 2.443164e-03 0.9671262 0.029 0.059 1.000000e+00 2
## RAB11FIP3 2.443315e-03 1.3158402 0.059 0.101 1.000000e+00 2
## RBBP9.1 2.460857e-03 0.8748522 0.012 0.034 1.000000e+00 2
## DGKE.1 2.469765e-03 1.4175631 0.068 0.114 1.000000e+00 2
## C2CD3.1 2.472203e-03 1.1938238 0.053 0.092 1.000000e+00 2
## PAIP2B 2.474341e-03 -1.2914483 0.003 0.019 1.000000e+00 2
## RGPD8 2.474416e-03 0.8711637 0.015 0.039 1.000000e+00 2
## NSMCE1-DT.1 2.475032e-03 0.5460380 0.005 0.022 1.000000e+00 2
## VSIG10 2.475032e-03 0.4540426 0.005 0.022 1.000000e+00 2
## AC114760.2 2.476656e-03 -0.3518295 0.005 0.022 1.000000e+00 2
## SULF2 2.476943e-03 -1.1944917 0.003 0.019 1.000000e+00 2
## RNF24.1 2.495345e-03 1.2857882 0.065 0.109 1.000000e+00 2
## ZNF566 2.496044e-03 0.3963294 0.009 0.030 1.000000e+00 2
## C11orf49.1 2.497932e-03 1.2632783 0.093 0.150 1.000000e+00 2
## ALG11 2.503391e-03 1.2756704 0.038 0.071 1.000000e+00 2
## RRAGB 2.505446e-03 1.4748802 0.026 0.054 1.000000e+00 2
## AC067852.2 2.520111e-03 1.0452838 0.020 0.046 1.000000e+00 2
## TRAF6 2.522311e-03 1.2054154 0.039 0.073 1.000000e+00 2
## NUP35.2 2.526268e-03 0.7067321 0.024 0.052 1.000000e+00 2
## GGT1.1 2.532765e-03 -1.0339272 0.002 0.017 1.000000e+00 2
## HIST1H4D.1 2.534236e-03 0.2974011 0.008 0.027 1.000000e+00 2
## PCGF3 2.536412e-03 1.2067634 0.027 0.056 1.000000e+00 2
## MFSD13A 2.540226e-03 0.9961192 0.008 0.027 1.000000e+00 2
## SPATA20 2.544215e-03 0.8587651 0.018 0.043 1.000000e+00 2
## MAK 2.544547e-03 0.8711632 0.011 0.032 1.000000e+00 2
## MESP1.1 2.548439e-03 -3.4135488 0.000 0.014 1.000000e+00 2
## ZNF121 2.555810e-03 1.3423837 0.050 0.088 1.000000e+00 2
## ARID3A 2.563363e-03 1.1754147 0.033 0.065 1.000000e+00 2
## NBPF10 2.580766e-03 1.1611299 0.030 0.061 1.000000e+00 2
## KLHL6.1 2.587979e-03 0.9244038 0.054 0.094 1.000000e+00 2
## SMYD5.2 2.600300e-03 -0.3062186 0.009 0.029 1.000000e+00 2
## AGBL5 2.601140e-03 0.6967622 0.012 0.034 1.000000e+00 2
## STRN 2.607617e-03 1.1235804 0.123 0.194 1.000000e+00 2
## UBE2O 2.610302e-03 1.4069471 0.066 0.111 1.000000e+00 2
## NAT1.1 2.611014e-03 1.3313803 0.015 0.039 1.000000e+00 2
## XRRA1.1 2.613376e-03 1.2913190 0.087 0.141 1.000000e+00 2
## ERMARD 2.624963e-03 0.9484642 0.044 0.080 1.000000e+00 2
## SP140 2.630387e-03 1.0127373 0.286 0.467 1.000000e+00 2
## SREBF2.1 2.632020e-03 1.3979251 0.120 0.189 1.000000e+00 2
## EME1.2 2.638744e-03 0.2854811 0.005 0.022 1.000000e+00 2
## PSRC1.2 2.647945e-03 -0.7751765 0.003 0.019 1.000000e+00 2
## DCUN1D4 2.655745e-03 1.0118885 0.033 0.065 1.000000e+00 2
## AC011446.2 2.655949e-03 0.6726151 0.009 0.029 1.000000e+00 2
## PPM1B 2.658600e-03 1.3896825 0.149 0.233 1.000000e+00 2
## EXD2.1 2.660016e-03 -0.1562100 0.003 0.019 1.000000e+00 2
## CRYBG3.1 2.660047e-03 0.8511654 0.032 0.063 1.000000e+00 2
## EDDM13.1 2.662644e-03 0.3486318 0.006 0.024 1.000000e+00 2
## C5orf58 2.667960e-03 0.5306553 0.009 0.029 1.000000e+00 2
## GPR19.1 2.667960e-03 0.9732643 0.009 0.029 1.000000e+00 2
## FAR2.2 2.672730e-03 1.2794550 0.074 0.122 1.000000e+00 2
## ZBTB21 2.674799e-03 0.9809380 0.032 0.063 1.000000e+00 2
## PYROXD1.1 2.677921e-03 1.2794541 0.087 0.141 1.000000e+00 2
## LMBR1 2.688321e-03 1.3551849 0.143 0.224 1.000000e+00 2
## TAF5 2.691952e-03 0.7984765 0.026 0.054 1.000000e+00 2
## KDM4C 2.692241e-03 1.8417009 0.301 0.333 1.000000e+00 2
## MTMR4.1 2.701130e-03 1.3713229 0.072 0.119 1.000000e+00 2
## EYA3.1 2.706030e-03 1.3610029 0.083 0.134 1.000000e+00 2
## ZCCHC4 2.707201e-03 1.2125551 0.035 0.067 1.000000e+00 2
## ZBTB17 2.712349e-03 1.4922787 0.041 0.075 1.000000e+00 2
## NDRG1 2.731157e-03 1.3646023 0.069 0.115 1.000000e+00 2
## FCRLB 2.735922e-03 -4.0488781 0.000 0.013 1.000000e+00 2
## AC005229.4 2.735922e-03 -3.8404218 0.000 0.013 1.000000e+00 2
## AC005695.2 2.735922e-03 -3.8280944 0.000 0.013 1.000000e+00 2
## B4GALT4 2.740004e-03 0.9672611 0.039 0.073 1.000000e+00 2
## STK38.1 2.743270e-03 1.0322104 0.242 0.382 1.000000e+00 2
## PLEKHB1 2.744688e-03 0.4707917 0.011 0.032 1.000000e+00 2
## FGFR1OP.1 2.752942e-03 1.1352004 0.057 0.098 1.000000e+00 2
## DEPDC5 2.753975e-03 1.2607313 0.033 0.065 1.000000e+00 2
## AC008083.2.1 2.755089e-03 0.5237076 0.014 0.036 1.000000e+00 2
## AC074032.1 2.776600e-03 1.0773983 0.026 0.054 1.000000e+00 2
## SYTL3.1 2.779469e-03 1.0228680 0.295 0.480 1.000000e+00 2
## STIM1.1 2.779849e-03 1.7545306 0.298 0.326 1.000000e+00 2
## IFT88.1 2.792246e-03 1.1832805 0.051 0.090 1.000000e+00 2
## TRIT1 2.800546e-03 0.8056622 0.032 0.062 1.000000e+00 2
## TPCN1.2 2.804081e-03 1.1987701 0.050 0.088 1.000000e+00 2
## DNAJC24.1 2.807549e-03 1.0552455 0.039 0.073 1.000000e+00 2
## AL359220.1.1 2.844722e-03 1.0304390 0.036 0.069 1.000000e+00 2
## ZNF829 2.848653e-03 0.2225804 0.003 0.019 1.000000e+00 2
## MORC2.2 2.849493e-03 1.1796153 0.042 0.077 1.000000e+00 2
## CCNT2 2.858610e-03 1.1102357 0.117 0.184 1.000000e+00 2
## ROCK1.1 2.865451e-03 0.9414160 0.305 0.500 1.000000e+00 2
## AC073332.1 2.865582e-03 1.7477043 0.050 0.088 1.000000e+00 2
## IPP 2.873961e-03 1.2285801 0.033 0.064 1.000000e+00 2
## TRAV5 2.884102e-03 -1.4627054 0.008 0.026 1.000000e+00 2
## ZNF333 2.891330e-03 0.2979960 0.014 0.036 1.000000e+00 2
## POLR1A.1 2.898168e-03 1.3738732 0.069 0.115 1.000000e+00 2
## AC099343.3 2.898479e-03 1.5062089 0.048 0.086 1.000000e+00 2
## KLHL5.2 2.902235e-03 1.3579732 0.116 0.182 1.000000e+00 2
## FBXO32 2.905074e-03 1.0516792 0.072 0.119 1.000000e+00 2
## ZNF816 2.910183e-03 0.8833692 0.011 0.031 1.000000e+00 2
## AC159540.2 2.910499e-03 -1.8960412 0.002 0.016 1.000000e+00 2
## C17orf97 2.911603e-03 -1.7786140 0.002 0.016 1.000000e+00 2
## AC008105.1 2.913812e-03 -1.8487564 0.002 0.016 1.000000e+00 2
## AC091982.3 2.914918e-03 -1.2570116 0.002 0.016 1.000000e+00 2
## CEP162.1 2.959476e-03 1.2518073 0.035 0.067 1.000000e+00 2
## PAQR3.1 2.984157e-03 1.1029568 0.032 0.062 1.000000e+00 2
## DCBLD1 2.992579e-03 0.8569935 0.021 0.047 1.000000e+00 2
## SRD5A1.1 3.003285e-03 1.5884061 0.029 0.058 1.000000e+00 2
## ARL17A 3.009903e-03 0.8149578 0.024 0.051 1.000000e+00 2
## ABHD2.1 3.010985e-03 1.4041751 0.080 0.130 1.000000e+00 2
## AP001267.1 3.021114e-03 1.0975180 0.026 0.054 1.000000e+00 2
## TRIM25.1 3.023249e-03 1.1815520 0.060 0.102 1.000000e+00 2
## ATP8B1 3.026116e-03 0.6628617 0.009 0.029 1.000000e+00 2
## AL137856.1 3.037230e-03 0.7924694 0.023 0.049 1.000000e+00 2
## NOCT.1 3.038214e-03 1.0820861 0.036 0.068 1.000000e+00 2
## ZNF492.2 3.041126e-03 -0.9788017 0.003 0.019 1.000000e+00 2
## CHCHD6.2 3.060939e-03 1.4276877 0.033 0.064 1.000000e+00 2
## IQCE.1 3.080385e-03 0.9816725 0.020 0.045 1.000000e+00 2
## TRGV2.2 3.082871e-03 0.4680757 0.012 0.034 1.000000e+00 2
## PAK4 3.093270e-03 0.9488522 0.018 0.043 1.000000e+00 2
## FAM149B1 3.097281e-03 0.9768765 0.027 0.056 1.000000e+00 2
## SIN3A.2 3.102314e-03 1.3182675 0.128 0.199 1.000000e+00 2
## MTMR12 3.123700e-03 1.4895753 0.050 0.087 1.000000e+00 2
## ZNF354C 3.123715e-03 -1.9763369 0.002 0.016 1.000000e+00 2
## AC010969.2 3.127270e-03 -1.3668300 0.002 0.016 1.000000e+00 2
## MACROD2 3.127270e-03 0.3741346 0.002 0.016 1.000000e+00 2
## CELF1 3.131086e-03 1.2087490 0.153 0.238 1.000000e+00 2
## AC068888.1.1 3.136225e-03 0.9486922 0.012 0.034 1.000000e+00 2
## ZNF585B 3.137502e-03 0.7350770 0.014 0.036 1.000000e+00 2
## ANKRD36B.1 3.144547e-03 1.4780431 0.129 0.202 1.000000e+00 2
## HPS4 3.145224e-03 1.2786292 0.057 0.098 1.000000e+00 2
## ZNF791 3.153486e-03 1.3059188 0.068 0.112 1.000000e+00 2
## ELF3-AS1 3.153792e-03 -3.8156146 0.000 0.013 1.000000e+00 2
## F8A1 3.153792e-03 -3.7204881 0.000 0.013 1.000000e+00 2
## CACNB2 3.153792e-03 -3.8685991 0.000 0.013 1.000000e+00 2
## TNFSF13B.2 3.174222e-03 0.9363718 0.027 0.055 1.000000e+00 2
## WNT5B 3.178407e-03 2.6599500 0.024 0.011 1.000000e+00 2
## LMBRD2 3.191848e-03 1.3053790 0.057 0.098 1.000000e+00 2
## FRMD4B.1 3.196091e-03 1.3218948 0.093 0.148 1.000000e+00 2
## PISD 3.219936e-03 1.3308014 0.062 0.104 1.000000e+00 2
## PPM1F.1 3.222992e-03 0.5430738 0.009 0.029 1.000000e+00 2
## KCNG2 3.228036e-03 -0.3228571 0.005 0.021 1.000000e+00 2
## OAF 3.238570e-03 0.1441900 0.005 0.021 1.000000e+00 2
## TRGC1.1 3.239625e-03 -0.2817778 0.005 0.021 1.000000e+00 2
## ATG2A 3.243034e-03 1.5069683 0.053 0.091 1.000000e+00 2
## COG6 3.247144e-03 1.3283856 0.066 0.110 1.000000e+00 2
## PTGIR.2 3.262547e-03 -0.6630221 0.003 0.019 1.000000e+00 2
## DYRK3 3.265961e-03 -0.2948011 0.003 0.019 1.000000e+00 2
## NRSN2-AS1 3.265961e-03 -0.5040187 0.003 0.019 1.000000e+00 2
## LMTK2.1 3.272711e-03 1.2751991 0.063 0.106 1.000000e+00 2
## IL10RA.2 3.285770e-03 0.8479683 0.350 0.593 1.000000e+00 2
## CCDC102B 3.291558e-03 -0.4770456 0.006 0.024 1.000000e+00 2
## TMEM25.2 3.299002e-03 0.7822022 0.008 0.026 1.000000e+00 2
## TMEM53 3.307415e-03 0.9092700 0.011 0.031 1.000000e+00 2
## AEBP2 3.321643e-03 1.3192925 0.177 0.277 1.000000e+00 2
## MFSD6 3.326594e-03 1.2312461 0.149 0.231 1.000000e+00 2
## PBX3.2 3.327277e-03 1.3183021 0.078 0.127 1.000000e+00 2
## BTBD2 3.328756e-03 1.3573632 0.045 0.081 1.000000e+00 2
## MINK1.1 3.340794e-03 1.4265599 0.098 0.155 1.000000e+00 2
## LPIN2 3.355021e-03 1.0508324 0.257 0.412 1.000000e+00 2
## AL445524.1 3.355218e-03 -0.5088274 0.002 0.016 1.000000e+00 2
## PIK3CA.1 3.360275e-03 1.1458645 0.086 0.137 1.000000e+00 2
## ADAM12 3.360736e-03 2.6987110 0.014 0.005 1.000000e+00 2
## AC005037.1.1 3.373494e-03 1.1038530 0.021 0.047 1.000000e+00 2
## MAVS 3.377891e-03 1.2291956 0.054 0.093 1.000000e+00 2
## STARD4.1 3.378904e-03 1.1895628 0.036 0.068 1.000000e+00 2
## AC068724.3 3.383812e-03 1.0590070 0.018 0.042 1.000000e+00 2
## PODXL2 3.386373e-03 -3.7477393 0.000 0.013 1.000000e+00 2
## CXCL16 3.386373e-03 -3.9103424 0.000 0.013 1.000000e+00 2
## RNF130.1 3.386924e-03 1.1850348 0.018 0.042 1.000000e+00 2
## MBLAC1 3.398019e-03 -0.4940466 0.005 0.021 1.000000e+00 2
## TESMIN.1 3.413340e-03 0.9169918 0.009 0.029 1.000000e+00 2
## AL139807.1.1 3.413678e-03 1.3529506 0.057 0.097 1.000000e+00 2
## DNAJC27-AS1 3.413957e-03 0.4815043 0.020 0.044 1.000000e+00 2
## POLM 3.419277e-03 1.3060211 0.059 0.099 1.000000e+00 2
## GCLC 3.419413e-03 1.2011884 0.063 0.106 1.000000e+00 2
## FBXO3 3.421109e-03 1.1979966 0.074 0.121 1.000000e+00 2
## GPR146 3.434376e-03 0.7466422 0.009 0.029 1.000000e+00 2
## EPHA4.2 3.439176e-03 1.1399019 0.030 0.059 1.000000e+00 2
## TRBV30.2 3.440249e-03 -1.9381972 0.009 0.028 1.000000e+00 2
## NPHP4.2 3.441626e-03 0.8941910 0.026 0.053 1.000000e+00 2
## PHOSPHO2 3.447041e-03 -0.5086859 0.005 0.021 1.000000e+00 2
## TULP4 3.451601e-03 1.2455576 0.078 0.127 1.000000e+00 2
## DCAF1.2 3.454386e-03 1.3708348 0.071 0.116 1.000000e+00 2
## BICD1.1 3.456321e-03 1.2505515 0.078 0.127 1.000000e+00 2
## SEMA4B 3.461399e-03 0.3351795 0.014 0.035 1.000000e+00 2
## AP000640.2 3.465433e-03 3.7485499 0.012 0.004 1.000000e+00 2
## EPC1.1 3.468256e-03 0.9618916 0.307 0.507 1.000000e+00 2
## WDR5B 3.470526e-03 -0.1322922 0.008 0.026 1.000000e+00 2
## SH3PXD2A.1 3.471535e-03 1.2902678 0.023 0.049 1.000000e+00 2
## TMPRSS6 3.474473e-03 3.5976235 0.012 0.004 1.000000e+00 2
## TNFRSF14-AS1 3.474481e-03 1.3437084 0.032 0.062 1.000000e+00 2
## C12orf4.1 3.479741e-03 1.3312098 0.053 0.091 1.000000e+00 2
## SEPSECS-AS1 3.480674e-03 0.3057544 0.008 0.026 1.000000e+00 2
## SPON1-AS1 3.481266e-03 3.5126048 0.012 0.004 1.000000e+00 2
## EARS2.1 3.482995e-03 0.7834184 0.017 0.040 1.000000e+00 2
## AC092919.1 3.484299e-03 -0.9136881 0.003 0.018 1.000000e+00 2
## ISM1.1 3.492804e-03 -0.2086854 0.003 0.018 1.000000e+00 2
## AC087276.2 3.502548e-03 -0.7168333 0.003 0.018 1.000000e+00 2
## LINC01806 3.514634e-03 0.6406968 0.014 0.035 1.000000e+00 2
## AC016065.1 3.515567e-03 0.7938423 0.011 0.031 1.000000e+00 2
## MAPK1.2 3.516495e-03 1.2135533 0.214 0.337 1.000000e+00 2
## LATS1 3.520219e-03 1.4104929 0.134 0.207 1.000000e+00 2
## PTCD2 3.522551e-03 0.6099925 0.014 0.035 1.000000e+00 2
## ZNF548 3.529004e-03 0.7630571 0.024 0.051 1.000000e+00 2
## TRUB1.1 3.531141e-03 1.2180517 0.030 0.059 1.000000e+00 2
## IPCEF1 3.532586e-03 1.1435148 0.057 0.097 1.000000e+00 2
## C5orf24 3.554270e-03 1.2208646 0.047 0.082 1.000000e+00 2
## DNM2 3.587240e-03 1.3145246 0.233 0.370 1.000000e+00 2
## SLC12A7.1 3.588081e-03 1.3810789 0.083 0.133 1.000000e+00 2
## SOS2 3.590663e-03 1.1209575 0.096 0.152 1.000000e+00 2
## PACSIN2.1 3.591159e-03 1.4917101 0.074 0.120 1.000000e+00 2
## ATN1.2 3.593778e-03 1.4863920 0.026 0.053 1.000000e+00 2
## MKRN2OS 3.598558e-03 -1.0526553 0.002 0.016 1.000000e+00 2
## PLEKHG4 3.598558e-03 -0.5987953 0.002 0.016 1.000000e+00 2
## GTPBP2.1 3.600611e-03 1.1938227 0.021 0.047 1.000000e+00 2
## METRNL.1 3.606844e-03 0.7294095 0.017 0.040 1.000000e+00 2
## PAXIP1-AS1 3.607693e-03 0.6689212 0.017 0.040 1.000000e+00 2
## FAM78A 3.612704e-03 1.3141216 0.075 0.122 1.000000e+00 2
## JAK3.1 3.631286e-03 1.3605210 0.220 0.347 1.000000e+00 2
## ZFP69 3.636322e-03 -3.4119173 0.000 0.013 1.000000e+00 2
## RAB30 3.636660e-03 0.9080083 0.030 0.059 1.000000e+00 2
## SLC30A6.1 3.637027e-03 1.1865597 0.069 0.114 1.000000e+00 2
## WARS2-AS1 3.639548e-03 1.3264758 0.053 0.091 1.000000e+00 2
## KDM4A 3.640738e-03 1.1702196 0.074 0.120 1.000000e+00 2
## ZNF583 3.647167e-03 0.4257968 0.009 0.028 1.000000e+00 2
## TSTD3 3.651290e-03 1.5361846 0.042 0.076 1.000000e+00 2
## SPATA2L 3.670087e-03 1.1645129 0.020 0.044 1.000000e+00 2
## MARK4 3.670754e-03 1.3069763 0.045 0.080 1.000000e+00 2
## CNKSR2.2 3.675276e-03 0.5583208 0.015 0.038 1.000000e+00 2
## ENO3 3.679832e-03 1.3776681 0.030 0.059 1.000000e+00 2
## FADS2.1 3.701244e-03 0.4850932 0.005 0.021 1.000000e+00 2
## OSBPL8 3.703017e-03 1.0070717 0.314 0.523 1.000000e+00 2
## CCR1.2 3.706053e-03 0.1221528 0.005 0.021 1.000000e+00 2
## DBT 3.719924e-03 0.8529618 0.029 0.057 1.000000e+00 2
## AC118549.1 3.723493e-03 1.2714550 0.122 0.189 1.000000e+00 2
## FCRL6.1 3.731620e-03 -0.1656416 0.008 0.026 1.000000e+00 2
## ZNF850 3.732707e-03 0.5452556 0.008 0.026 1.000000e+00 2
## TNIP3.1 3.733793e-03 0.1650352 0.008 0.026 1.000000e+00 2
## SRSF11 3.734235e-03 0.8387374 0.365 0.619 1.000000e+00 2
## AC104316.1 3.734555e-03 0.7395608 0.011 0.031 1.000000e+00 2
## RNF144A-AS1.2 3.751079e-03 -0.7188892 0.003 0.018 1.000000e+00 2
## ADIPOR2 3.756505e-03 1.2503386 0.111 0.174 1.000000e+00 2
## RNF31.1 3.800983e-03 1.2412768 0.062 0.103 1.000000e+00 2
## LINC01970 3.809111e-03 3.2057622 0.024 0.011 1.000000e+00 2
## PRR12 3.828948e-03 1.4798995 0.048 0.084 1.000000e+00 2
## SLC9A6.1 3.834006e-03 1.0997135 0.026 0.053 1.000000e+00 2
## ST3GAL5.2 3.864681e-03 1.1136571 0.059 0.099 1.000000e+00 2
## ABHD8.1 3.879298e-03 1.4301488 0.020 0.044 1.000000e+00 2
## AC009690.2 3.888038e-03 0.7006742 0.009 0.028 1.000000e+00 2
## HIP1R 3.888898e-03 1.0835018 0.045 0.080 1.000000e+00 2
## ARHGAP11B.2 3.890686e-03 0.9062116 0.030 0.059 1.000000e+00 2
## ATP2A2.2 3.893154e-03 1.4084546 0.120 0.187 1.000000e+00 2
## GPR35.1 3.898040e-03 1.3974589 0.017 0.040 1.000000e+00 2
## SYCP2.1 3.904361e-03 1.2848588 0.015 0.037 1.000000e+00 2
## CCDC163 3.904962e-03 -3.6103387 0.000 0.012 1.000000e+00 2
## C1orf53 3.904962e-03 -3.4512981 0.000 0.012 1.000000e+00 2
## TCEAL3 3.904962e-03 -4.1571438 0.000 0.012 1.000000e+00 2
## ENTPD7 3.905007e-03 0.3122235 0.008 0.026 1.000000e+00 2
## SLC44A1.2 3.932018e-03 0.1714794 0.014 0.035 1.000000e+00 2
## TLK2.1 3.947370e-03 1.4199068 0.105 0.164 1.000000e+00 2
## RP2 3.951623e-03 1.0706884 0.036 0.067 1.000000e+00 2
## VPS13C 3.952408e-03 1.5721325 0.371 0.467 1.000000e+00 2
## ZNF324B 3.953184e-03 0.6384818 0.011 0.030 1.000000e+00 2
## SOCS7 3.953184e-03 0.5770771 0.011 0.030 1.000000e+00 2
## CEP192.2 3.976539e-03 1.1479348 0.152 0.233 1.000000e+00 2
## SLC31A2.1 3.989897e-03 0.3997598 0.008 0.026 1.000000e+00 2
## ZNF484 4.002962e-03 1.1766914 0.039 0.072 1.000000e+00 2
## EAF2.2 4.020630e-03 1.4415283 0.035 0.065 1.000000e+00 2
## MDFIC.1 4.024215e-03 1.3031436 0.454 0.627 1.000000e+00 2
## MIR222HG.1 4.043045e-03 2.5756443 0.045 0.026 1.000000e+00 2
## LINC00649.1 4.048094e-03 1.3244941 0.132 0.204 1.000000e+00 2
## AC087294.1 4.060368e-03 3.3412221 0.015 0.006 1.000000e+00 2
## SYNRG.1 4.068272e-03 1.6695341 0.349 0.419 1.000000e+00 2
## MKNK1.2 4.095294e-03 1.3545249 0.101 0.158 1.000000e+00 2
## ASH1L 4.101164e-03 1.4948249 0.376 0.468 1.000000e+00 2
## MLYCD 4.103517e-03 1.3669306 0.035 0.065 1.000000e+00 2
## UBXN7 4.104061e-03 1.4643740 0.072 0.118 1.000000e+00 2
## DCP1A 4.117178e-03 1.0660993 0.111 0.172 1.000000e+00 2
## ZNF736.2 4.132247e-03 0.9904808 0.029 0.057 1.000000e+00 2
## SNX32 4.136487e-03 0.9587279 0.009 0.028 1.000000e+00 2
## DMC1.2 4.136841e-03 -1.5670140 0.002 0.015 1.000000e+00 2
## PEX11A 4.144677e-03 -0.5481263 0.002 0.015 1.000000e+00 2
## ZNF335 4.157977e-03 1.5767277 0.087 0.138 1.000000e+00 2
## CNNM3.1 4.164473e-03 1.2243745 0.053 0.090 1.000000e+00 2
## FEZ2.1 4.166093e-03 1.2154764 0.056 0.094 1.000000e+00 2
## ANGEL2 4.169542e-03 1.3029597 0.083 0.132 1.000000e+00 2
## PDK3.1 4.190503e-03 1.2823806 0.104 0.162 1.000000e+00 2
## AC010976.1.1 4.193717e-03 -3.6389260 0.000 0.012 1.000000e+00 2
## POLR3G 4.193717e-03 -3.4881261 0.000 0.012 1.000000e+00 2
## TOLLIP-AS1.2 4.193717e-03 -3.9280121 0.000 0.012 1.000000e+00 2
## ZNF219 4.193717e-03 -3.9844683 0.000 0.012 1.000000e+00 2
## AC004158.1 4.193717e-03 -3.3786787 0.000 0.012 1.000000e+00 2
## CLSTN1 4.203741e-03 1.3358533 0.147 0.228 1.000000e+00 2
## AP000873.2 4.208522e-03 0.6337885 0.012 0.033 1.000000e+00 2
## KAT14.1 4.208946e-03 0.7849905 0.011 0.030 1.000000e+00 2
## CDKL1.1 4.226571e-03 1.2362086 0.041 0.073 1.000000e+00 2
## ZNF514 4.230149e-03 0.6886540 0.012 0.033 1.000000e+00 2
## HSPA14 4.243582e-03 0.2238348 0.008 0.026 1.000000e+00 2
## BBS2 4.254627e-03 1.3529808 0.032 0.061 1.000000e+00 2
## NICN1 4.269511e-03 0.9154020 0.015 0.037 1.000000e+00 2
## IKZF4.1 4.270003e-03 0.5939071 0.020 0.044 1.000000e+00 2
## TSNARE1.1 4.270686e-03 1.4398687 0.032 0.061 1.000000e+00 2
## MAP1A 4.272384e-03 0.7182445 0.017 0.039 1.000000e+00 2
## AC058791.1 4.280062e-03 0.7525851 0.024 0.050 1.000000e+00 2
## AC009126.1 4.292469e-03 0.9214551 0.024 0.050 1.000000e+00 2
## SLC26A4-AS1.1 4.306924e-03 0.6577675 0.003 0.018 1.000000e+00 2
## UFSP1 4.308418e-03 -0.2581752 0.003 0.018 1.000000e+00 2
## MAN1B1-DT 4.308418e-03 -0.2487767 0.003 0.018 1.000000e+00 2
## AC015849.1.1 4.315176e-03 2.4255815 0.120 0.095 1.000000e+00 2
## GPR89B 4.318365e-03 1.2992756 0.036 0.067 1.000000e+00 2
## ADNP2.2 4.338494e-03 1.3446457 0.042 0.075 1.000000e+00 2
## JHY.1 4.356344e-03 0.2321403 0.009 0.028 1.000000e+00 2
## KHNYN 4.356941e-03 1.4853019 0.059 0.099 1.000000e+00 2
## ZMIZ1 4.361253e-03 1.3641889 0.075 0.121 1.000000e+00 2
## CENPO.2 4.363031e-03 1.2813537 0.026 0.052 1.000000e+00 2
## STAM.1 4.375839e-03 1.2529758 0.105 0.164 1.000000e+00 2
## AC092652.1 4.382690e-03 0.7972359 0.015 0.037 1.000000e+00 2
## SMG7.1 4.383300e-03 1.2666665 0.170 0.262 1.000000e+00 2
## DUSP5.1 4.406636e-03 1.2140262 0.090 0.142 1.000000e+00 2
## BTBD7 4.409214e-03 1.4634463 0.092 0.145 1.000000e+00 2
## MXD1 4.426882e-03 1.4734399 0.090 0.142 1.000000e+00 2
## IQCH 4.437430e-03 -1.5706690 0.002 0.015 1.000000e+00 2
## RAD9B 4.445833e-03 -1.0017594 0.002 0.015 1.000000e+00 2
## MYO9B 4.451507e-03 1.8276278 0.322 0.371 1.000000e+00 2
## GGA3 4.463925e-03 1.2778171 0.065 0.107 1.000000e+00 2
## SLC16A7.1 4.468305e-03 1.2737140 0.198 0.308 1.000000e+00 2
## MFSD9 4.486821e-03 0.8228971 0.011 0.030 1.000000e+00 2
## IL6R.2 4.497584e-03 0.1736608 0.005 0.020 1.000000e+00 2
## TTC38.1 4.501242e-03 0.8794764 0.020 0.043 1.000000e+00 2
## NDN.2 4.504124e-03 -3.7216177 0.000 0.012 1.000000e+00 2
## DERPC 4.504124e-03 -3.4913100 0.000 0.012 1.000000e+00 2
## COPZ2 4.504124e-03 -3.5225545 0.000 0.012 1.000000e+00 2
## AC021092.1 4.504855e-03 0.3984413 0.005 0.020 1.000000e+00 2
## AL162253.2 4.510015e-03 1.3082609 0.033 0.063 1.000000e+00 2
## HEATR5A 4.513632e-03 1.2667033 0.024 0.050 1.000000e+00 2
## BMERB1 4.515018e-03 1.2364850 0.032 0.061 1.000000e+00 2
## PAXBP1.2 4.520627e-03 1.5300033 0.149 0.229 1.000000e+00 2
## POLR2J2 4.561007e-03 1.0933186 0.030 0.058 1.000000e+00 2
## MTERF2 4.587089e-03 -0.1355506 0.006 0.023 1.000000e+00 2
## LNX2 4.591137e-03 0.8835886 0.032 0.060 1.000000e+00 2
## NIBAN2 4.599677e-03 1.2124749 0.023 0.048 1.000000e+00 2
## TAF5L 4.600927e-03 1.2436793 0.045 0.079 1.000000e+00 2
## ZNF707 4.605562e-03 -0.2612141 0.009 0.028 1.000000e+00 2
## TAF4B.2 4.605639e-03 0.9752013 0.041 0.073 1.000000e+00 2
## KIF27 4.606933e-03 1.3432177 0.030 0.058 1.000000e+00 2
## ARHGAP35.1 4.609902e-03 2.1967786 0.159 0.139 1.000000e+00 2
## C2orf81 4.611206e-03 0.4978097 0.015 0.037 1.000000e+00 2
## PDCD1LG2 4.612688e-03 -0.8287644 0.003 0.018 1.000000e+00 2
## INTS6-AS1 4.617067e-03 1.2830272 0.021 0.046 1.000000e+00 2
## AC022916.1 4.645373e-03 1.0625845 0.029 0.056 1.000000e+00 2
## RAP1GAP2.1 4.652831e-03 1.4283761 0.108 0.168 1.000000e+00 2
## NAA25.2 4.663199e-03 1.4070613 0.083 0.132 1.000000e+00 2
## CEP164 4.666477e-03 1.3392044 0.063 0.104 1.000000e+00 2
## ATP7A 4.716469e-03 0.8017647 0.029 0.056 1.000000e+00 2
## SPATA5.2 4.716826e-03 1.5651915 0.099 0.155 1.000000e+00 2
## TDRKH 4.733779e-03 0.8835058 0.018 0.041 1.000000e+00 2
## ZNF180 4.737045e-03 0.8436344 0.023 0.048 1.000000e+00 2
## IFI44.2 4.751949e-03 1.3668436 0.033 0.063 1.000000e+00 2
## CLOCK 4.752724e-03 1.4523459 0.057 0.096 1.000000e+00 2
## TAF1A 4.760173e-03 0.4237315 0.012 0.032 1.000000e+00 2
## ZNF90.2 4.769085e-03 -1.3975384 0.002 0.015 1.000000e+00 2
## AL121944.1 4.772693e-03 -0.5382842 0.002 0.015 1.000000e+00 2
## TCAF2 4.804638e-03 0.9245846 0.036 0.067 1.000000e+00 2
## BX664615.2 4.841439e-03 0.8714014 0.008 0.025 1.000000e+00 2
## MTF1 4.841988e-03 1.2908824 0.045 0.079 1.000000e+00 2
## B3GAT2 4.849124e-03 -0.1572547 0.005 0.020 1.000000e+00 2
## AL691403.1.1 4.858230e-03 1.5143256 0.078 0.125 1.000000e+00 2
## DIAPH2.2 4.884398e-03 1.9982568 0.236 0.237 1.000000e+00 2
## PIGV 4.891312e-03 1.2330023 0.042 0.075 1.000000e+00 2
## ZNFX1 4.898733e-03 1.2031722 0.188 0.289 1.000000e+00 2
## PPME1.1 4.938362e-03 1.3838026 0.069 0.112 1.000000e+00 2
## TSGA10 4.954919e-03 1.1025715 0.023 0.047 1.000000e+00 2
## ERICH6-AS1 4.979004e-03 1.0633496 0.015 0.037 1.000000e+00 2
## TRAV1-2.1 4.998978e-03 -0.8313025 0.009 0.027 1.000000e+00 2
## CEP70 5.009459e-03 0.8474532 0.018 0.041 1.000000e+00 2
## GLCE.2 5.010992e-03 0.7498373 0.014 0.034 1.000000e+00 2
## TARSL2 5.021531e-03 1.1824812 0.090 0.142 1.000000e+00 2
## TRDV1 5.029490e-03 -1.4268700 0.006 0.022 1.000000e+00 2
## SYNGR3 5.034612e-03 0.3644756 0.011 0.030 1.000000e+00 2
## KDM3B.1 5.044105e-03 1.2568944 0.125 0.192 1.000000e+00 2
## AP002381.2 5.066851e-03 1.0251396 0.017 0.039 1.000000e+00 2
## Z93930.2 5.077923e-03 1.4170947 0.074 0.118 1.000000e+00 2
## RPRD1A.1 5.097760e-03 1.2425658 0.084 0.133 1.000000e+00 2
## KTI12 5.121884e-03 0.4135464 0.002 0.015 1.000000e+00 2
## IPPK 5.150107e-03 1.1501320 0.036 0.066 1.000000e+00 2
## POGLUT3 5.154072e-03 0.7502150 0.027 0.054 1.000000e+00 2
## MDM4 5.162508e-03 1.2147470 0.161 0.246 1.000000e+00 2
## GPR137B.1 5.176602e-03 1.1313360 0.041 0.072 1.000000e+00 2
## FAM131A 5.181012e-03 -0.1786872 0.005 0.020 1.000000e+00 2
## FKBP1B.1 5.196656e-03 -3.7828116 0.000 0.012 1.000000e+00 2
## POC1B.2 5.202587e-03 1.5135670 0.111 0.172 1.000000e+00 2
## GALNT11.1 5.269064e-03 1.2834320 0.080 0.127 1.000000e+00 2
## TRMT10A 5.269908e-03 0.7032588 0.014 0.034 1.000000e+00 2
## THBS4 5.283776e-03 -1.1684890 0.003 0.017 1.000000e+00 2
## AC073569.2 5.294741e-03 -0.2013567 0.003 0.017 1.000000e+00 2
## ZNF784 5.298400e-03 0.3897045 0.003 0.017 1.000000e+00 2
## WDR91 5.312694e-03 0.7071765 0.020 0.043 1.000000e+00 2
## CYB5D2 5.326512e-03 1.4595690 0.041 0.072 1.000000e+00 2
## PUS10 5.335010e-03 1.1415204 0.026 0.051 1.000000e+00 2
## B3GALNT2.1 5.369510e-03 1.1755559 0.027 0.054 1.000000e+00 2
## UTY 5.396343e-03 1.7350726 0.310 0.354 1.000000e+00 2
## ATXN7L1.1 5.409912e-03 0.8975564 0.041 0.072 1.000000e+00 2
## SLC2A1 5.420531e-03 1.4087146 0.078 0.124 1.000000e+00 2
## APH1B 5.443942e-03 1.1760504 0.032 0.060 1.000000e+00 2
## CLIC4.2 5.457070e-03 0.9962460 0.018 0.041 1.000000e+00 2
## AC010260.1.1 5.460667e-03 -0.3271896 0.008 0.025 1.000000e+00 2
## MNT.1 5.465036e-03 1.5824403 0.033 0.062 1.000000e+00 2
## NOTCH2NLC 5.467253e-03 2.3441449 0.098 0.074 1.000000e+00 2
## IKBKB.2 5.472431e-03 1.4932845 0.113 0.174 1.000000e+00 2
## NACC2 5.473559e-03 1.3332049 0.029 0.055 1.000000e+00 2
## PLCB3.1 5.479462e-03 0.7873227 0.015 0.036 1.000000e+00 2
## LINC02802 5.496887e-03 -1.1064080 0.002 0.014 1.000000e+00 2
## AMIGO1.1 5.496887e-03 -1.5434191 0.002 0.014 1.000000e+00 2
## MTUS1 5.496887e-03 -1.5369585 0.002 0.014 1.000000e+00 2
## USP12-AS1 5.496887e-03 -0.7428312 0.002 0.014 1.000000e+00 2
## PLLP 5.496887e-03 -1.0557281 0.002 0.014 1.000000e+00 2
## SLC35A3.1 5.503754e-03 1.1276602 0.065 0.105 1.000000e+00 2
## NIPA1 5.523058e-03 1.0600919 0.036 0.066 1.000000e+00 2
## AC245060.5 5.528375e-03 0.8326196 0.023 0.047 1.000000e+00 2
## PIGA 5.553825e-03 1.2714332 0.035 0.064 1.000000e+00 2
## AP002812.2 5.563380e-03 0.2605143 0.006 0.022 1.000000e+00 2
## TRAM2-AS1 5.582499e-03 -3.7027491 0.000 0.011 1.000000e+00 2
## FGF14-AS2 5.582499e-03 -3.4598609 0.000 0.011 1.000000e+00 2
## LKAAEAR1.2 5.582499e-03 -3.4147957 0.000 0.011 1.000000e+00 2
## SLC29A2 5.585269e-03 0.5433127 0.006 0.022 1.000000e+00 2
## AC010149.1 5.596758e-03 1.2112615 0.018 0.041 1.000000e+00 2
## LINC01765.1 5.614029e-03 0.8152122 0.020 0.043 1.000000e+00 2
## TTC21B-AS1 5.643525e-03 -1.4667712 0.003 0.017 1.000000e+00 2
## UBE2W 5.665112e-03 1.2037608 0.105 0.163 1.000000e+00 2
## TRIM68 5.670182e-03 1.1211017 0.011 0.029 1.000000e+00 2
## RNF43 5.670854e-03 -1.0233319 0.003 0.017 1.000000e+00 2
## AL031728.1 5.670854e-03 -0.1856470 0.003 0.017 1.000000e+00 2
## TRBV6-2 5.672456e-03 -2.2582028 0.005 0.020 1.000000e+00 2
## DDX43 5.672811e-03 -0.4106359 0.003 0.017 1.000000e+00 2
## VWA5A 5.672811e-03 -0.5130007 0.003 0.017 1.000000e+00 2
## EXOC3-AS1 5.685074e-03 0.3066114 0.011 0.029 1.000000e+00 2
## AC026202.2 5.691009e-03 0.8773968 0.012 0.032 1.000000e+00 2
## TNFRSF10D 5.693884e-03 0.7888594 0.012 0.032 1.000000e+00 2
## VPS52 5.694210e-03 1.4969787 0.059 0.097 1.000000e+00 2
## RPTOR 5.721716e-03 1.9208756 0.250 0.258 1.000000e+00 2
## MRTFB.1 5.724514e-03 1.4259456 0.041 0.072 1.000000e+00 2
## NLRP1.1 5.750612e-03 1.3493529 0.168 0.257 1.000000e+00 2
## SNRK.1 5.759185e-03 1.0978168 0.179 0.273 1.000000e+00 2
## AP2B1 5.786944e-03 1.1999841 0.174 0.268 1.000000e+00 2
## ELMOD3.1 5.789870e-03 1.1603596 0.039 0.070 1.000000e+00 2
## ZFY 5.795598e-03 1.3509953 0.068 0.109 1.000000e+00 2
## AC005670.3.1 5.811267e-03 1.2778134 0.084 0.132 1.000000e+00 2
## RUNDC1 5.823328e-03 1.0556748 0.023 0.047 1.000000e+00 2
## TRAV24.1 5.827313e-03 -0.6241996 0.006 0.022 1.000000e+00 2
## GTDC1 5.833936e-03 2.1948282 0.173 0.156 1.000000e+00 2
## ABR.1 5.842700e-03 2.2944721 0.167 0.149 1.000000e+00 2
## FBXL22 5.856135e-03 0.2461365 0.009 0.027 1.000000e+00 2
## GATD3A 5.861584e-03 0.6966755 0.008 0.025 1.000000e+00 2
## MLF1 5.863287e-03 -0.8240985 0.005 0.020 1.000000e+00 2
## TIPARP-AS1 5.866614e-03 0.3094881 0.008 0.025 1.000000e+00 2
## PIGL 5.880599e-03 1.4845483 0.084 0.132 1.000000e+00 2
## AC027644.3 5.885117e-03 1.1148749 0.026 0.051 1.000000e+00 2
## LINC01089.1 5.889157e-03 0.8088737 0.018 0.040 1.000000e+00 2
## CFLAR.2 5.908669e-03 1.2207208 0.236 0.371 1.000000e+00 2
## NIPBL-DT 5.922766e-03 1.1180396 0.020 0.043 1.000000e+00 2
## HIST1H2AJ.1 5.931973e-03 0.1946135 0.006 0.022 1.000000e+00 2
## ZNF382 5.946294e-03 0.6825578 0.006 0.022 1.000000e+00 2
## VGLL4 5.963157e-03 1.3659939 0.113 0.173 1.000000e+00 2
## SLFN13 5.965827e-03 0.7983441 0.033 0.061 1.000000e+00 2
## AC004596.1 5.971431e-03 0.7037467 0.006 0.022 1.000000e+00 2
## RASGRP3.1 5.975030e-03 0.2200863 0.006 0.022 1.000000e+00 2
## EHBP1L1.1 5.997252e-03 1.5341751 0.090 0.141 1.000000e+00 2
## FAM92A 5.997455e-03 -3.4216150 0.000 0.011 1.000000e+00 2
## RLN2.1 5.997455e-03 -3.9007886 0.000 0.011 1.000000e+00 2
## RBM26-AS1 5.997455e-03 -3.6146359 0.000 0.011 1.000000e+00 2
## ZNF284 5.997455e-03 -3.7662623 0.000 0.011 1.000000e+00 2
## BTG3-AS1 5.997455e-03 -3.8276326 0.000 0.011 1.000000e+00 2
## CD274 5.998531e-03 0.4625500 0.009 0.027 1.000000e+00 2
## ZNF91 6.020614e-03 1.2266811 0.107 0.164 1.000000e+00 2
## DST.2 6.064107e-03 0.9439044 0.018 0.040 1.000000e+00 2
## AC036176.1.1 6.071680e-03 0.2755793 0.003 0.017 1.000000e+00 2
## ZSCAN18 6.071680e-03 -0.6143781 0.003 0.017 1.000000e+00 2
## NEK3 6.082147e-03 0.1469398 0.003 0.017 1.000000e+00 2
## BBS4 6.082156e-03 0.9995507 0.029 0.055 1.000000e+00 2
## BCAS3 6.182568e-03 2.0811871 0.226 0.224 1.000000e+00 2
## ACVR2A.1 6.200892e-03 2.5786230 0.093 0.070 1.000000e+00 2
## PPARG 6.202909e-03 3.9797212 0.014 0.005 1.000000e+00 2
## TTC33.1 6.217696e-03 1.3984861 0.110 0.168 1.000000e+00 2
## APLF.1 6.227567e-03 0.7148949 0.020 0.042 1.000000e+00 2
## PIAS1 6.241921e-03 1.2489962 0.215 0.335 1.000000e+00 2
## MRNIP 6.244209e-03 1.1886487 0.033 0.061 1.000000e+00 2
## RTTN.2 6.256292e-03 0.9744562 0.059 0.096 1.000000e+00 2
## STK35.2 6.262004e-03 0.7388004 0.026 0.051 1.000000e+00 2
## METTL22 6.264577e-03 1.4732469 0.032 0.059 1.000000e+00 2
## CYFIP1.2 6.286161e-03 1.3823042 0.033 0.061 1.000000e+00 2
## AP4S1 6.308148e-03 1.3335421 0.027 0.053 1.000000e+00 2
## KLF9 6.319234e-03 1.3037502 0.077 0.121 1.000000e+00 2
## AP001318.2 6.325060e-03 -1.1187583 0.002 0.014 1.000000e+00 2
## KDM7A-DT 6.329836e-03 -1.2229249 0.002 0.014 1.000000e+00 2
## CXorf21 6.330937e-03 -0.3486742 0.005 0.019 1.000000e+00 2
## FNBP4 6.334956e-03 1.2631297 0.281 0.454 1.000000e+00 2
## ADCK5 6.339159e-03 1.1855154 0.029 0.055 1.000000e+00 2
## P3H4 6.343095e-03 0.1214381 0.005 0.019 1.000000e+00 2
## PINLYP 6.370571e-03 0.6042157 0.006 0.022 1.000000e+00 2
## ZC3H12D 6.379125e-03 1.7765223 0.053 0.088 1.000000e+00 2
## ACSF3.1 6.392679e-03 1.3279148 0.098 0.151 1.000000e+00 2
## AFAP1.2 6.399737e-03 0.6958289 0.009 0.027 1.000000e+00 2
## TIMD4.1 6.443772e-03 -4.0807487 0.000 0.011 1.000000e+00 2
## FXYD1 6.443772e-03 -3.9459357 0.000 0.011 1.000000e+00 2
## AC097382.2 6.443772e-03 -3.6195386 0.000 0.011 1.000000e+00 2
## LRRC8C.1 6.445204e-03 1.2871363 0.162 0.247 1.000000e+00 2
## TIRAP 6.452515e-03 0.7968582 0.023 0.047 1.000000e+00 2
## USP3 6.461935e-03 1.2223876 0.230 0.361 1.000000e+00 2
## DHX57 6.462916e-03 1.2068250 0.036 0.065 1.000000e+00 2
## CEP250 6.471197e-03 1.2668165 0.126 0.192 1.000000e+00 2
## SAT1 6.500445e-03 1.2973901 0.311 0.500 1.000000e+00 2
## C19orf38.2 6.507654e-03 -0.6139137 0.003 0.017 1.000000e+00 2
## AC011447.3.2 6.547570e-03 0.2812349 0.008 0.024 1.000000e+00 2
## ETV3 6.562497e-03 1.2755480 0.084 0.131 1.000000e+00 2
## MIR3142HG.1 6.581224e-03 0.1496570 0.008 0.024 1.000000e+00 2
## CBWD1 6.585138e-03 1.1973229 0.015 0.036 1.000000e+00 2
## ZFAND4.1 6.609531e-03 1.1034124 0.029 0.055 1.000000e+00 2
## TRGV8.1 6.616916e-03 0.3727282 0.008 0.024 1.000000e+00 2
## ZC3H10 6.626338e-03 0.4600080 0.008 0.024 1.000000e+00 2
## THRA 6.631350e-03 1.1274917 0.020 0.042 1.000000e+00 2
## AL645728.1.1 6.662966e-03 0.7554905 0.017 0.038 1.000000e+00 2
## SUN1.2 6.668532e-03 1.2059188 0.045 0.078 1.000000e+00 2
## HINFP 6.697894e-03 1.0913869 0.036 0.065 1.000000e+00 2
## ACSL4 6.726978e-03 1.4249180 0.122 0.186 1.000000e+00 2
## TUT7.1 6.737714e-03 1.5466898 0.161 0.246 1.000000e+00 2
## ACSF2 6.755244e-03 1.2450553 0.017 0.038 1.000000e+00 2
## KPTN.1 6.777384e-03 1.0179735 0.009 0.026 1.000000e+00 2
## HIST1H4F 6.795785e-03 0.9540293 0.009 0.026 1.000000e+00 2
## FRMD5.2 6.796894e-03 -0.1160693 0.002 0.014 1.000000e+00 2
## LRRC75A 6.796894e-03 -0.6880313 0.002 0.014 1.000000e+00 2
## BDNF-AS 6.802247e-03 0.1916585 0.006 0.022 1.000000e+00 2
## LINC01260 6.810408e-03 0.7649024 0.006 0.022 1.000000e+00 2
## POMGNT2 6.810408e-03 0.4404424 0.006 0.022 1.000000e+00 2
## TMEM260 6.835928e-03 1.3004494 0.078 0.123 1.000000e+00 2
## KIF5C.1 6.850289e-03 0.4381476 0.053 0.087 1.000000e+00 2
## MTHFD2L 6.877345e-03 1.0469569 0.033 0.061 1.000000e+00 2
## ZNF737.1 6.909617e-03 1.0616741 0.041 0.071 1.000000e+00 2
## SIRPG-AS1 6.923881e-03 -3.6426474 0.000 0.011 1.000000e+00 2
## DDX31 6.962911e-03 1.4441796 0.033 0.061 1.000000e+00 2
## LGALS8-AS1 6.973954e-03 4.2557818 0.015 0.006 1.000000e+00 2
## TCTN1 6.977423e-03 0.1439498 0.003 0.017 1.000000e+00 2
## HYPK 6.977423e-03 -0.7572492 0.003 0.017 1.000000e+00 2
## TRBV12-4.1 6.979603e-03 -1.4587962 0.015 0.035 1.000000e+00 2
## B3GNT10 6.982217e-03 0.2723644 0.003 0.017 1.000000e+00 2
## RBL2.2 6.995150e-03 0.8999447 0.347 0.576 1.000000e+00 2
## ABCC4.1 7.042176e-03 1.3946414 0.056 0.092 1.000000e+00 2
## NSD1 7.043907e-03 1.1099585 0.191 0.291 1.000000e+00 2
## MAFF 7.056713e-03 0.6773466 0.015 0.035 1.000000e+00 2
## TRBV4-1.1 7.057572e-03 -1.8746113 0.014 0.032 1.000000e+00 2
## SIRT1 7.072518e-03 1.0893196 0.063 0.102 1.000000e+00 2
## GCNA 7.077161e-03 0.9403305 0.014 0.033 1.000000e+00 2
## TBC1D25 7.089323e-03 1.2349104 0.053 0.088 1.000000e+00 2
## ZNF318 7.093451e-03 1.1841301 0.020 0.042 1.000000e+00 2
## TATDN2 7.094580e-03 1.4547596 0.083 0.129 1.000000e+00 2
## MYO5A.1 7.128103e-03 2.0145506 0.236 0.241 1.000000e+00 2
## HOMER2.1 7.135240e-03 1.1602589 0.017 0.038 1.000000e+00 2
## SMAD5 7.135729e-03 1.2809961 0.029 0.055 1.000000e+00 2
## TLN1.1 7.136451e-03 0.8672813 0.361 0.616 1.000000e+00 2
## TMEM39A 7.151109e-03 1.1983963 0.089 0.137 1.000000e+00 2
## TOP3B 7.201518e-03 1.1362602 0.030 0.057 1.000000e+00 2
## NKIRAS1 7.208088e-03 0.8721719 0.009 0.026 1.000000e+00 2
## CUL1.2 7.224493e-03 1.2651080 0.179 0.272 1.000000e+00 2
## AC020913.1.1 7.226301e-03 -0.1470068 0.005 0.019 1.000000e+00 2
## BBOF1 7.235513e-03 0.1306198 0.005 0.019 1.000000e+00 2
## LINC02580 7.251279e-03 0.8779337 0.012 0.031 1.000000e+00 2
## LINC00092 7.256344e-03 0.1275312 0.006 0.022 1.000000e+00 2
## AL139099.1 7.258589e-03 0.9197571 0.005 0.019 1.000000e+00 2
## LINC00891.1 7.296063e-03 -0.2439614 0.002 0.014 1.000000e+00 2
## AK9 7.321598e-03 0.9834735 0.035 0.063 1.000000e+00 2
## R3HCC1L 7.323325e-03 1.3503496 0.116 0.176 1.000000e+00 2
## PHF8.1 7.325115e-03 1.2928370 0.063 0.102 1.000000e+00 2
## L3MBTL3 7.334470e-03 1.0546415 0.045 0.077 1.000000e+00 2
## ZNF551 7.347476e-03 1.3876878 0.015 0.035 1.000000e+00 2
## TRAF3IP2-AS1 7.347658e-03 1.4167606 0.044 0.075 1.000000e+00 2
## GATA3-AS1.2 7.357751e-03 1.1392150 0.015 0.035 1.000000e+00 2
## QKI.1 7.360475e-03 1.4380706 0.174 0.266 1.000000e+00 2
## SSH2.1 7.375918e-03 1.5902620 0.370 0.468 1.000000e+00 2
## AC008124.1 7.390827e-03 1.1538338 0.026 0.050 1.000000e+00 2
## SLC39A13 7.407101e-03 0.9853713 0.042 0.073 1.000000e+00 2
## ARHGAP26.1 7.410319e-03 2.0356741 0.227 0.228 1.000000e+00 2
## ZNF510 7.410917e-03 0.7238485 0.017 0.037 1.000000e+00 2
## CD79A 7.432598e-03 -1.3265229 0.003 0.016 1.000000e+00 2
## AL121787.1 7.440407e-03 -3.6121503 0.000 0.011 1.000000e+00 2
## AC090061.1 7.440407e-03 -3.8431985 0.000 0.011 1.000000e+00 2
## TBXA2R.1 7.440407e-03 -3.9851861 0.000 0.011 1.000000e+00 2
## PHETA2 7.440407e-03 -3.4077146 0.000 0.011 1.000000e+00 2
## FNTB.1 7.442459e-03 1.0334418 0.035 0.063 1.000000e+00 2
## CHST10 7.442806e-03 0.1277914 0.003 0.016 1.000000e+00 2
## PIGZ.1 7.476071e-03 -0.2486883 0.003 0.016 1.000000e+00 2
## ELK3 7.485648e-03 1.2416432 0.053 0.087 1.000000e+00 2
## MAPK8 7.499008e-03 1.2627551 0.123 0.186 1.000000e+00 2
## ANKS6 7.512793e-03 0.6445942 0.008 0.024 1.000000e+00 2
## MAP3K14-AS1 7.520338e-03 1.1839894 0.021 0.044 1.000000e+00 2
## PKN3.2 7.542615e-03 0.8214487 0.008 0.024 1.000000e+00 2
## SRCAP 7.545811e-03 1.3816915 0.093 0.144 1.000000e+00 2
## RTL10 7.566977e-03 0.2493202 0.009 0.026 1.000000e+00 2
## PDE8A.2 7.576589e-03 1.3583431 0.063 0.102 1.000000e+00 2
## ZNF43.2 7.582136e-03 1.7259137 0.048 0.081 1.000000e+00 2
## MYSM1 7.588356e-03 1.3845943 0.107 0.164 1.000000e+00 2
## AC108134.3 7.606937e-03 0.9432635 0.033 0.060 1.000000e+00 2
## LINC00662 7.611503e-03 1.5684226 0.033 0.061 1.000000e+00 2
## AC018647.3 7.620568e-03 3.2884385 0.011 0.003 1.000000e+00 2
## C16orf71 7.630309e-03 3.6795959 0.011 0.003 1.000000e+00 2
## ITGAM.2 7.634932e-03 0.9428777 0.048 0.081 1.000000e+00 2
## LDLR.2 7.647644e-03 1.3121213 0.107 0.162 1.000000e+00 2
## HSPBAP1 7.649139e-03 0.9675313 0.030 0.056 1.000000e+00 2
## CDC37L1-DT 7.663535e-03 0.4434301 0.009 0.026 1.000000e+00 2
## CCDC91 7.673016e-03 1.3575523 0.218 0.338 1.000000e+00 2
## PTPRE.1 7.687888e-03 1.5125289 0.068 0.108 1.000000e+00 2
## ZNF468 7.705084e-03 0.9497950 0.020 0.042 1.000000e+00 2
## AARS2 7.706933e-03 0.5859985 0.021 0.044 1.000000e+00 2
## FHOD1 7.715320e-03 1.6206149 0.071 0.112 1.000000e+00 2
## SLC48A1 7.715345e-03 0.9237152 0.009 0.026 1.000000e+00 2
## HCG11 7.727849e-03 1.2685131 0.011 0.028 1.000000e+00 2
## SMC1B.1 7.754672e-03 0.7899041 0.005 0.019 1.000000e+00 2
## WAKMAR2.1 7.775668e-03 1.3094443 0.051 0.085 1.000000e+00 2
## AL355574.1 7.817596e-03 -1.8223664 0.002 0.014 1.000000e+00 2
## TIGD7 7.826439e-03 -1.4599110 0.002 0.014 1.000000e+00 2
## CCDC7 7.836034e-03 1.3843831 0.120 0.182 1.000000e+00 2
## ARMH4 7.847373e-03 1.2199268 0.032 0.058 1.000000e+00 2
## PXYLP1 7.925085e-03 1.3760257 0.041 0.071 1.000000e+00 2
## ZNF768 7.996187e-03 -3.2201590 0.000 0.010 1.000000e+00 2
## SLC5A10 7.996187e-03 -3.4200815 0.000 0.010 1.000000e+00 2
## PSMA8 7.996187e-03 -3.6215080 0.000 0.010 1.000000e+00 2
## ACTR5 7.999126e-03 1.4075340 0.023 0.046 1.000000e+00 2
## RNF111 8.003180e-03 1.2698710 0.140 0.210 1.000000e+00 2
## SPIN3 8.013224e-03 -0.5275524 0.003 0.016 1.000000e+00 2
## C18orf54.1 8.013224e-03 0.7381634 0.003 0.016 1.000000e+00 2
## SLC7A5.1 8.027595e-03 2.1894151 0.168 0.152 1.000000e+00 2
## CSGALNACT1.2 8.033187e-03 1.1545503 0.069 0.109 1.000000e+00 2
## MMP24 8.046478e-03 0.3111187 0.012 0.030 1.000000e+00 2
## MBOAT1.1 8.054230e-03 1.3980704 0.057 0.093 1.000000e+00 2
## ZCWPW2 8.054656e-03 1.3083773 0.035 0.062 1.000000e+00 2
## ACTR3B 8.056071e-03 0.5920070 0.018 0.039 1.000000e+00 2
## AL135925.1 8.084551e-03 1.7558850 0.024 0.048 1.000000e+00 2
## PEX7.1 8.095988e-03 1.0155461 0.020 0.041 1.000000e+00 2
## NME7.1 8.105589e-03 1.4253186 0.105 0.160 1.000000e+00 2
## VPS50 8.110397e-03 1.3348374 0.087 0.135 1.000000e+00 2
## TCF25 8.116110e-03 1.3355343 0.260 0.413 1.000000e+00 2
## CAMTA2.1 8.122299e-03 1.4338198 0.050 0.083 1.000000e+00 2
## TRBV3-1 8.140368e-03 -1.1421366 0.012 0.030 1.000000e+00 2
## FGL2 8.165188e-03 0.5440967 0.011 0.028 1.000000e+00 2
## ERI2.2 8.191074e-03 1.0954850 0.009 0.026 1.000000e+00 2
## NFE2L1.1 8.228914e-03 1.2101463 0.035 0.062 1.000000e+00 2
## Z93241.1.1 8.268661e-03 2.2872744 0.138 0.117 1.000000e+00 2
## DZANK1 8.274042e-03 0.2772418 0.005 0.019 1.000000e+00 2
## PLA2G15.1 8.278833e-03 1.4054052 0.020 0.041 1.000000e+00 2
## NLK 8.285290e-03 2.4390150 0.110 0.087 1.000000e+00 2
## PPARD.1 8.302615e-03 1.2464325 0.033 0.060 1.000000e+00 2
## HDAC8 8.348696e-03 1.2874504 0.134 0.202 1.000000e+00 2
## IRF4.1 8.352705e-03 1.4142432 0.047 0.078 1.000000e+00 2
## ZNF354A 8.371130e-03 1.2847561 0.021 0.043 1.000000e+00 2
## SLC29A3 8.382659e-03 0.4366265 0.012 0.030 1.000000e+00 2
## CD160.1 8.389550e-03 -1.9277870 0.002 0.013 1.000000e+00 2
## CES1.1 8.405365e-03 -1.3854521 0.002 0.013 1.000000e+00 2
## AC080112.1 8.408531e-03 -0.3983235 0.002 0.013 1.000000e+00 2
## PDE9A 8.408531e-03 -0.3543495 0.002 0.013 1.000000e+00 2
## FKBP14.1 8.415935e-03 0.4817085 0.012 0.030 1.000000e+00 2
## TRAPPC8 8.423596e-03 1.5072346 0.116 0.175 1.000000e+00 2
## ZNF786 8.441601e-03 1.2146610 0.026 0.050 1.000000e+00 2
## PTGDR.1 8.442901e-03 0.9241886 0.024 0.048 1.000000e+00 2
## AC016727.1 8.451137e-03 0.8853652 0.014 0.033 1.000000e+00 2
## MTERF1 8.536486e-03 1.4572965 0.065 0.103 1.000000e+00 2
## AC073342.1 8.553931e-03 -0.5359006 0.003 0.016 1.000000e+00 2
## SLC3A1 8.583230e-03 -0.5245592 0.003 0.016 1.000000e+00 2
## PARS2 8.589101e-03 0.3545964 0.003 0.016 1.000000e+00 2
## AC107081.2 8.594289e-03 -3.2384624 0.000 0.010 1.000000e+00 2
## LINC00239.1 8.594289e-03 -2.9009883 0.000 0.010 1.000000e+00 2
## CBX2.1 8.594289e-03 -3.3571910 0.000 0.010 1.000000e+00 2
## SIGLEC10 8.594289e-03 -3.4389857 0.000 0.010 1.000000e+00 2
## LENG9 8.594699e-03 1.0327199 0.012 0.030 1.000000e+00 2
## GRAMD4.1 8.604785e-03 0.7839270 0.015 0.035 1.000000e+00 2
## SLC25A44 8.669243e-03 1.5309722 0.039 0.068 1.000000e+00 2
## GANC 8.690155e-03 0.8882375 0.029 0.054 1.000000e+00 2
## CEACAM1.1 8.702667e-03 1.3514521 0.035 0.062 1.000000e+00 2
## CYP2U1 8.715562e-03 -0.3023127 0.006 0.021 1.000000e+00 2
## LYSMD4 8.715812e-03 1.1395282 0.011 0.028 1.000000e+00 2
## TBCD.2 8.726958e-03 1.3083392 0.189 0.286 1.000000e+00 2
## ST8SIA6.1 8.769383e-03 0.7505763 0.023 0.045 1.000000e+00 2
## METTL25 8.782741e-03 1.4996180 0.059 0.095 1.000000e+00 2
## SLC2A8.1 8.797751e-03 0.1862046 0.014 0.032 1.000000e+00 2
## HDAC4-AS1 8.801407e-03 0.1491917 0.006 0.021 1.000000e+00 2
## ZNF669 8.825189e-03 1.0493439 0.024 0.047 1.000000e+00 2
## KCNK12.1 8.839299e-03 -0.4865357 0.005 0.018 1.000000e+00 2
## BBS12 8.858898e-03 -0.3445459 0.005 0.018 1.000000e+00 2
## MYOM2 8.900174e-03 0.6704839 0.015 0.034 1.000000e+00 2
## EXOC6B.2 8.901936e-03 2.1244381 0.192 0.182 1.000000e+00 2
## DHX35 8.902053e-03 1.5249251 0.024 0.047 1.000000e+00 2
## BAIAP2L1 8.953184e-03 -2.2789849 0.002 0.013 1.000000e+00 2
## DNAJC18 8.956205e-03 0.9964373 0.014 0.032 1.000000e+00 2
## ACACA.1 8.967925e-03 1.2332968 0.072 0.113 1.000000e+00 2
## ZNF441 8.972428e-03 0.2483121 0.009 0.026 1.000000e+00 2
## MLLT3.1 8.973511e-03 1.2588530 0.227 0.352 1.000000e+00 2
## LPIN1 8.984409e-03 1.3215536 0.134 0.201 1.000000e+00 2
## ETNK1 9.002584e-03 1.3889633 0.189 0.290 1.000000e+00 2
## AC018797.3 9.020871e-03 -1.5933849 0.002 0.013 1.000000e+00 2
## MIATNB 9.021374e-03 1.3948356 0.063 0.101 1.000000e+00 2
## METTL7A.1 9.024267e-03 -1.6787884 0.002 0.013 1.000000e+00 2
## NDRG2.1 9.024267e-03 -0.4249067 0.002 0.013 1.000000e+00 2
## CTDSPL.2 9.025966e-03 -0.5474447 0.002 0.013 1.000000e+00 2
## INO80 9.027410e-03 1.6076383 0.135 0.204 1.000000e+00 2
## CD44 9.043033e-03 0.8120621 0.380 0.633 1.000000e+00 2
## ARHGEF9 9.051620e-03 1.5381003 0.081 0.126 1.000000e+00 2
## YTHDF3 9.055066e-03 1.5128647 0.111 0.168 1.000000e+00 2
## RBM44 9.061803e-03 2.8571961 0.015 0.006 1.000000e+00 2
## CBWD3.1 9.112675e-03 -1.6744794 0.003 0.016 1.000000e+00 2
## CALCB.1 9.127947e-03 0.5342179 0.011 0.028 1.000000e+00 2
## EVI5L 9.132043e-03 1.3859771 0.018 0.039 1.000000e+00 2
## EEF2K 9.146003e-03 1.3574158 0.059 0.095 1.000000e+00 2
## DHTKD1.1 9.164102e-03 1.0966683 0.033 0.060 1.000000e+00 2
## ECM2.2 9.181403e-03 -0.4004498 0.003 0.016 1.000000e+00 2
## OMD.1 9.193948e-03 0.1180142 0.003 0.016 1.000000e+00 2
## ABCA3.1 9.202224e-03 1.1858533 0.015 0.034 1.000000e+00 2
## FAM185A 9.209410e-03 1.0255804 0.023 0.045 1.000000e+00 2
## SPON1.1 9.231866e-03 0.9095133 0.015 0.034 1.000000e+00 2
## JAKMIP1 9.238030e-03 -3.5427545 0.000 0.010 1.000000e+00 2
## TICAM2 9.238030e-03 -3.0871235 0.000 0.010 1.000000e+00 2
## CARD9 9.238030e-03 -3.5371567 0.000 0.010 1.000000e+00 2
## CCDC15-DT 9.238030e-03 -3.4763003 0.000 0.010 1.000000e+00 2
## EP400.1 9.258334e-03 1.2684981 0.141 0.212 1.000000e+00 2
## ZNF25 9.270103e-03 1.1997777 0.015 0.034 1.000000e+00 2
## TDRD3 9.277459e-03 1.3128990 0.095 0.144 1.000000e+00 2
## DDX17 9.344162e-03 1.2162446 0.453 0.627 1.000000e+00 2
## LIF.2 9.378583e-03 1.1360579 0.014 0.032 1.000000e+00 2
## SLC31A1.1 9.390406e-03 1.4292906 0.024 0.047 1.000000e+00 2
## TSC2 9.397301e-03 1.6935136 0.045 0.076 1.000000e+00 2
## ASNS 9.487858e-03 0.8350048 0.005 0.018 1.000000e+00 2
## AC008467.1 9.488946e-03 2.9680045 0.017 0.007 1.000000e+00 2
## ITGA1.2 9.541181e-03 2.3297132 0.081 0.060 1.000000e+00 2
## GRAP2.1 9.554632e-03 1.4704091 0.394 0.515 1.000000e+00 2
## ZNF623 9.556784e-03 0.7315409 0.023 0.045 1.000000e+00 2
## MLLT6.1 9.579753e-03 1.2959796 0.128 0.191 1.000000e+00 2
## TTN-AS1.1 9.591561e-03 0.9959030 0.054 0.088 1.000000e+00 2
## RASA2.2 9.592312e-03 1.2429470 0.262 0.412 1.000000e+00 2
## PIK3C3 9.611686e-03 1.4625781 0.143 0.215 1.000000e+00 2
## AC027097.1 9.629788e-03 0.6053684 0.012 0.030 1.000000e+00 2
## FZD6.1 9.653271e-03 0.1431296 0.008 0.023 1.000000e+00 2
## RAB11FIP2 9.661416e-03 1.4713315 0.068 0.107 1.000000e+00 2
## DACT1.2 9.682068e-03 -1.6541260 0.002 0.013 1.000000e+00 2
## SYCP3.1 9.693001e-03 -0.5965159 0.002 0.013 1.000000e+00 2
## LINC01547 9.693001e-03 -1.0794628 0.002 0.013 1.000000e+00 2
## KLF13.2 9.703704e-03 0.8884880 0.349 0.577 1.000000e+00 2
## MTSS1 9.713436e-03 0.1401079 0.012 0.030 1.000000e+00 2
## TMEM168 9.728152e-03 1.2639569 0.060 0.096 1.000000e+00 2
## AC022075.1.2 9.734015e-03 1.0679633 0.047 0.078 1.000000e+00 2
## EXOC2 9.741860e-03 1.4165410 0.164 0.247 1.000000e+00 2
## CD8B2.1 9.791394e-03 -1.0913950 0.003 0.016 1.000000e+00 2
## IL4.1 9.798074e-03 -1.2808605 0.003 0.016 1.000000e+00 2
## AC072022.2 9.819061e-03 0.8443156 0.011 0.028 1.000000e+00 2
## RNASE6.1 9.821564e-03 0.9696447 0.011 0.028 1.000000e+00 2
## RNF32 9.826411e-03 0.8244790 0.009 0.025 1.000000e+00 2
## MMP25.1 9.828714e-03 1.1965159 0.161 0.239 1.000000e+00 2
## UNC119B 9.829028e-03 0.6978273 0.009 0.025 1.000000e+00 2
## SEL1L3.2 9.833355e-03 1.3402444 0.192 0.293 1.000000e+00 2
## ILDR2.2 9.861736e-03 0.5786041 0.003 0.016 1.000000e+00 2
## NEURL1.1 9.861736e-03 1.2831591 0.003 0.016 1.000000e+00 2
## PGM2L1 9.865565e-03 1.4161619 0.041 0.070 1.000000e+00 2
## AC021188.1.1 9.899954e-03 0.7606230 0.032 0.057 1.000000e+00 2
## PTPN4.2 9.916161e-03 1.0801096 0.275 0.434 1.000000e+00 2
## AGTPBP1 9.925657e-03 1.5344046 0.158 0.237 1.000000e+00 2
## ENPP4.1 9.925842e-03 1.2134141 0.035 0.062 1.000000e+00 2
## TP73.2 9.930997e-03 -3.4802974 0.000 0.010 1.000000e+00 2
## LRP5 9.930997e-03 -3.4740602 0.000 0.010 1.000000e+00 2
## AC023509.2 9.930997e-03 -3.4979579 0.000 0.010 1.000000e+00 2
## ANKRD52.1 9.948315e-03 1.3767955 0.045 0.076 1.000000e+00 2
## NNT.1 9.949993e-03 1.2194539 0.108 0.163 1.000000e+00 2
## AC064807.1.1 9.967903e-03 0.9348203 0.014 0.032 1.000000e+00 2
## PPP1R12C 9.973272e-03 1.3900379 0.062 0.098 1.000000e+00 2
## ACACB 9.976957e-03 0.2563264 0.012 0.030 1.000000e+00 2
## gene
## RPS4X.1 RPS4X
## RPL10.1 RPL10
## RPLP1.2 RPLP1
## RPS14.2 RPS14
## RPS7.1 RPS7
## RPL41.2 RPL41
## RPL26.2 RPL26
## RPS13.2 RPS13
## RPS3A.1 RPS3A
## RPL18A.1 RPL18A
## RPS3.1 RPS3
## RPL30.2 RPL30
## RPL18.1 RPL18
## EEF1A1.1 EEF1A1
## RPS15A.2 RPS15A
## RPS12.1 RPS12
## TMSB4X.2 TMSB4X
## RPS24.2 RPS24
## RPL19.1 RPL19
## RPL7A.1 RPL7A
## RPL11.1 RPL11
## B2M.2 B2M
## RPL14.1 RPL14
## RPS27A.2 RPS27A
## RPL32.1 RPL32
## RPL15.1 RPL15
## RPL29.1 RPL29
## RPS5.2 RPS5
## RPL3.1 RPL3
## RPL8.1 RPL8
## FAU.2 FAU
## RPL39.1 RPL39
## RPS28.1 RPS28
## RPL6.2 RPL6
## RPSA.2 RPSA
## RACK1.1 RACK1
## MT-CO1.1 MT-CO1
## RPL9.2 RPL9
## RPS6.2 RPS6
## RPL5.1 RPL5
## RPL10A.1 RPL10A
## RPS23.1 RPS23
## RPL34.1 RPL34
## RPS25.1 RPS25
## BTF3.1 BTF3
## MALAT1.1 MALAT1
## RPS2.2 RPS2
## RPS16.1 RPS16
## EEF1B2.2 EEF1B2
## RPLP0.2 RPLP0
## RPL23A.1 RPL23A
## NACA.2 NACA
## RPS8.2 RPS8
## RPL35A.1 RPL35A
## RPS9.1 RPS9
## SH3BGRL3.2 SH3BGRL3
## PFN1.1 PFN1
## RPS18.2 RPS18
## UBA52.2 UBA52
## RPL24.1 RPL24
## RPL28.1 RPL28
## MT-CO2.2 MT-CO2
## RPL21.2 RPL21
## PPIA.1 PPIA
## IFITM1.1 IFITM1
## COX4I1.2 COX4I1
## GAPDH.1 GAPDH
## TPT1.2 TPT1
## FTL.2 FTL
## RPL12.2 RPL12
## RPLP2.1 RPLP2
## RPL17.1 RPL17
## MT-CYB.2 MT-CYB
## MYL12A.2 MYL12A
## RPL22.1 RPL22
## RPL7.2 RPL7
## HINT1.2 HINT1
## ATP5MC2.2 ATP5MC2
## CLIC1.2 CLIC1
## EEF1G.2 EEF1G
## MT-CO3.2 MT-CO3
## RPL27.1 RPL27
## MT-ATP6.2 MT-ATP6
## RPL13.1 RPL13
## MYL6.1 MYL6
## ACTB.1 ACTB
## LDHB.2 LDHB
## RPL37.1 RPL37
## CD52.2 CD52
## EIF3K.2 EIF3K
## PFDN5.2 PFDN5
## ATP5MG.2 ATP5MG
## ARPC3.2 ARPC3
## IFITM2.2 IFITM2
## CFL1.1 CFL1
## COX7C.2 COX7C
## RPS15.1 RPS15
## RPS27.2 RPS27
## RPS21.1 RPS21
## NDUFA4.1 NDUFA4
## RPL36AL.2 RPL36AL
## PSME1.2 PSME1
## MYL12B.2 MYL12B
## RPS19.2 RPS19
## S100A4.2 S100A4
## RPS26.2 RPS26
## TMSB10.1 TMSB10
## OST4.2 OST4
## SNHG29.2 SNHG29
## CHCHD2 CHCHD2
## NME2.1 NME2
## RPS4Y1.2 RPS4Y1
## ACTG1.1 ACTG1
## NDUFB11.2 NDUFB11
## ATP5F1E.2 ATP5F1E
## RNASEK.2 RNASEK
## SSR4.2 SSR4
## COMMD6.2 COMMD6
## S100A10.2 S100A10
## FTH1.1 FTH1
## SERF2.2 SERF2
## RPS17.1 RPS17
## IL32.2 IL32
## TOMM7.2 TOMM7
## CD3D.2 CD3D
## RPL35.1 RPL35
## HLA-B.2 HLA-B
## SRP14.2 SRP14
## PPIB.2 PPIB
## S100A6.2 S100A6
## ATP5MC3.1 ATP5MC3
## OAZ1.2 OAZ1
## COX6A1.1 COX6A1
## FXYD5.1 FXYD5
## EMP3.2 EMP3
## GABARAP.2 GABARAP
## SLC25A5 SLC25A5
## HCST.2 HCST
## RPS29.1 RPS29
## CD3E.2 CD3E
## ATP5PO.2 ATP5PO
## HLA-A.2 HLA-A
## COX6B1.2 COX6B1
## RPL4.2 RPL4
## S100A11.2 S100A11
## RPL36.1 RPL36
## SEC61B.2 SEC61B
## COX6C.1 COX6C
## RPL36A.1 RPL36A
## HLA-C.2 HLA-C
## RPL37A.1 RPL37A
## TRAPPC1.2 TRAPPC1
## COX8A.1 COX8A
## NDUFA11.1 NDUFA11
## SUMO2.1 SUMO2
## HNRNPA1.2 HNRNPA1
## H3F3A.2 H3F3A
## PARK7.2 PARK7
## SNRPD2.2 SNRPD2
## RPL27A.1 RPL27A
## PPDPF.2 PPDPF
## RPL31.1 RPL31
## BRK1.2 BRK1
## LY6E.1 LY6E
## SLC25A6.2 SLC25A6
## RPS11.1 RPS11
## UQCRH.1 UQCRH
## HIGD2A.2 HIGD2A
## TMA7.2 TMA7
## RAC2.2 RAC2
## COX5A COX5A
## NEDD8.2 NEDD8
## PTMA.1 PTMA
## CUTA.2 CUTA
## UBB.2 UBB
## UQCRB.2 UQCRB
## ABRACL.2 ABRACL
## PRDX2 PRDX2
## PPP1CA.2 PPP1CA
## RPL23.1 RPL23
## SSR2.2 SSR2
## CSTB.1 CSTB
## APRT.2 APRT
## PSMB9.1 PSMB9
## COX7A2.2 COX7A2
## UQCR11.2 UQCR11
## PCBP1.2 PCBP1
## GMFG.2 GMFG
## EIF3F.2 EIF3F
## UXT.1 UXT
## MIF MIF
## PLAAT4.1 PLAAT4
## NDUFB9.1 NDUFB9
## C9orf16.2 C9orf16
## MT-ND4L.2 MT-ND4L
## PSMB1.2 PSMB1
## TOMM6.1 TOMM6
## EEF2.1 EEF2
## NPM1.1 NPM1
## PSMB8.2 PSMB8
## PSMA7.2 PSMA7
## EIF5A EIF5A
## UBL5.2 UBL5
## ATP5PD.2 ATP5PD
## TRMT112.2 TRMT112
## MTRNR2L12.2 MTRNR2L12
## MT-ND5.2 MT-ND5
## CD3G.2 CD3G
## TPI1 TPI1
## EDF1.2 EDF1
## SOD1 SOD1
## TAGLN2.2 TAGLN2
## ATP5PF.1 ATP5PF
## CD7.2 CD7
## VAMP8.2 VAMP8
## RBM3 RBM3
## PCED1B-AS1.2 PCED1B-AS1
## KRTCAP2.2 KRTCAP2
## SF3B5.2 SF3B5
## CDK2AP2.2 CDK2AP2
## GYPC.2 GYPC
## SPCS1.2 SPCS1
## RPS20 RPS20
## EIF3E.1 EIF3E
## MT-ND1.2 MT-ND1
## CRIP1.2 CRIP1
## ATP6V0E1.2 ATP6V0E1
## ERH.2 ERH
## SEC61G.2 SEC61G
## PSMB3.1 PSMB3
## NDUFB8.2 NDUFB8
## TLE5.2 TLE5
## ATP5PB.2 ATP5PB
## NDUFC2.1 NDUFC2
## RAN RAN
## GNG5.2 GNG5
## SUB1.2 SUB1
## ERP29.2 ERP29
## TIMP1.2 TIMP1
## ARF5.2 ARF5
## EIF3L.1 EIF3L
## ATP5MF.2 ATP5MF
## EIF3G.2 EIF3G
## DBI.2 DBI
## COX5B.1 COX5B
## RHOA.1 RHOA
## GPX4 GPX4
## NDUFS5.2 NDUFS5
## RABAC1.2 RABAC1
## FIS1.2 FIS1
## RPS27L.2 RPS27L
## CSNK2B.2 CSNK2B
## DYNLL1.1 DYNLL1
## ARHGDIB.2 ARHGDIB
## H2AFZ.1 H2AFZ
## WDR83OS.2 WDR83OS
## RSL1D1.1 RSL1D1
## CD48.2 CD48
## PSME2.1 PSME2
## NDUFA12.2 NDUFA12
## ARPC1B.2 ARPC1B
## UQCR10.1 UQCR10
## GUK1.2 GUK1
## CD99.2 CD99
## TALDO1.1 TALDO1
## RPL38.1 RPL38
## OSTC.2 OSTC
## NDUFB2.1 NDUFB2
## TRIR.2 TRIR
## TXN TXN
## PSMA2.1 PSMA2
## DAD1.1 DAD1
## YBX1.1 YBX1
## BST2.1 BST2
## PRR13.2 PRR13
## NDUFA13.1 NDUFA13
## CYBA.2 CYBA
## BAX.2 BAX
## C19orf53.2 C19orf53
## ATP5IF1.1 ATP5IF1
## PRDX5.1 PRDX5
## MZT2B.1 MZT2B
## NOP10.1 NOP10
## GPSM3.2 GPSM3
## PSMB6.1 PSMB6
## RPL13A.2 RPL13A
## ATP5F1B.2 ATP5F1B
## MDH2.2 MDH2
## ENO1.2 ENO1
## ATP5F1D.2 ATP5F1D
## C4orf3.2 C4orf3
## YWHAB.2 YWHAB
## CHCHD10.2 CHCHD10
## PHB2.1 PHB2
## PRELID1.1 PRELID1
## DAZAP2.2 DAZAP2
## LSM7.1 LSM7
## GNB2.2 GNB2
## HMGN2.1 HMGN2
## MT-ND4.2 MT-ND4
## BANF1 BANF1
## SELENOH SELENOH
## C1QBP.1 C1QBP
## NDUFA1.1 NDUFA1
## TAF10.2 TAF10
## SRP9.1 SRP9
## CD63.2 CD63
## UBE2L6.1 UBE2L6
## ANAPC11 ANAPC11
## ELOB.1 ELOB
## ATP6V1G1.2 ATP6V1G1
## RTRAF.2 RTRAF
## CIB1.2 CIB1
## EEF1D.2 EEF1D
## LAMTOR4.2 LAMTOR4
## GTF3A.1 GTF3A
## TSPO.2 TSPO
## LTB.2 LTB
## ATP5MD.1 ATP5MD
## DRAP1.2 DRAP1
## MZT2A.2 MZT2A
## SLC25A3.2 SLC25A3
## COX7A2L.2 COX7A2L
## SELENOW.1 SELENOW
## ANP32B.1 ANP32B
## GSTP1.1 GSTP1
## LAPTM5.2 LAPTM5
## SNU13.2 SNU13
## GAS5.1 GAS5
## TSTD1.2 TSTD1
## IL2RG.2 IL2RG
## UBE2I.2 UBE2I
## SMDT1.2 SMDT1
## RGS10.2 RGS10
## COX7B.1 COX7B
## PSMB10.1 PSMB10
## PSMA5.1 PSMA5
## POMP.1 POMP
## CCNI.2 CCNI
## ATP6V0B.1 ATP6V0B
## SNRPB SNRPB
## PSMD8.1 PSMD8
## C12orf57.2 C12orf57
## NDUFB10.2 NDUFB10
## SAP18.2 SAP18
## MRPL51.1 MRPL51
## ANAPC16.2 ANAPC16
## JTB.2 JTB
## UCP2.2 UCP2
## PEBP1 PEBP1
## CALM3.2 CALM3
## PRDX6.2 PRDX6
## ATP6V0C.2 ATP6V0C
## LGALS1.2 LGALS1
## EIF3I EIF3I
## ATP6V1F.2 ATP6V1F
## PGLS.2 PGLS
## DDOST.2 DDOST
## CNBP.2 CNBP
## TMEM258.2 TMEM258
## LDHA.1 LDHA
## NDUFAB1.1 NDUFAB1
## KRT10.2 KRT10
## ATP5F1A.2 ATP5F1A
## PGK1 PGK1
## FBL.1 FBL
## CD37.2 CD37
## MAL.2 MAL
## NOSIP.1 NOSIP
## SF3B6.2 SF3B6
## UBXN1.2 UBXN1
## NDUFS7.2 NDUFS7
## SOS1.1 SOS1
## TUBB.2 TUBB
## ICAM3.2 ICAM3
## UFC1.2 UFC1
## TOMM22.2 TOMM22
## CD27.2 CD27
## PYCARD.2 PYCARD
## PHPT1.1 PHPT1
## CIAO2B.1 CIAO2B
## GABARAPL2.2 GABARAPL2
## SNHG6.1 SNHG6
## SNRPC SNRPC
## IFITM3 IFITM3
## GIMAP4.2 GIMAP4
## ITM2B.2 ITM2B
## RASAL2.1 RASAL2
## FKBP1A.1 FKBP1A
## PGAM1.2 PGAM1
## ARPC4.1 ARPC4
## SIT1.1 SIT1
## SSBP1 SSBP1
## DDT.1 DDT
## POLR2L.2 POLR2L
## RAC1.1 RAC1
## NDUFB4 NDUFB4
## NDUFA2 NDUFA2
## ERGIC3.2 ERGIC3
## COPE.2 COPE
## OCIAD2.2 OCIAD2
## NHP2 NHP2
## EIF3H.1 EIF3H
## GZMM.2 GZMM
## RBX1.1 RBX1
## ATP5F1C.1 ATP5F1C
## CAPNS1.2 CAPNS1
## DCTN3.2 DCTN3
## LAMTOR5.1 LAMTOR5
## ARPC5.1 ARPC5
## HSPA8.1 HSPA8
## PPA1 PPA1
## SEC11C.2 SEC11C
## MRPS21.2 MRPS21
## TRBC2.2 TRBC2
## MICOS10.2 MICOS10
## C17orf49.2 C17orf49
## CD81.2 CD81
## ISCU.2 ISCU
## SNRPG.2 SNRPG
## ALKBH7.2 ALKBH7
## TBCA.1 TBCA
## UQCRFS1.2 UQCRFS1
## TUFM.2 TUFM
## NDUFA7.1 NDUFA7
## PSMA6.1 PSMA6
## VAMP5.1 VAMP5
## UQCRQ.1 UQCRQ
## TMEM50A.2 TMEM50A
## NSA2.1 NSA2
## CCT6A CCT6A
## PRDX1 PRDX1
## PLP2.2 PLP2
## SRSF9.1 SRSF9
## BCAP31.1 BCAP31
## TWF2.2 TWF2
## NDUFS8.1 NDUFS8
## CORO1A.2 CORO1A
## GNAI2.2 GNAI2
## TXNL4A.2 TXNL4A
## SSR3.1 SSR3
## CNN2.2 CNN2
## SNX3.1 SNX3
## HSPE1 HSPE1
## NDUFB3.1 NDUFB3
## SERP1.1 SERP1
## ARRB2.2 ARRB2
## CALM1.2 CALM1
## TBCB.1 TBCB
## UBE2L3.1 UBE2L3
## NOP53.1 NOP53
## LMAN2.2 LMAN2
## VPS28.2 VPS28
## PPP4C.1 PPP4C
## GPI.2 GPI
## CST7.2 CST7
## SDHC.1 SDHC
## GHITM.1 GHITM
## HMGN1 HMGN1
## ARL6IP4.2 ARL6IP4
## TOMM20.2 TOMM20
## ARPC2.2 ARPC2
## MDH1.1 MDH1
## ST13 ST13
## SF3B4.1 SF3B4
## SEC11A.2 SEC11A
## LSM2 LSM2
## SEM1.1 SEM1
## GZMA.1 GZMA
## RSL24D1.2 RSL24D1
## COTL1.2 COTL1
## MGST3.1 MGST3
## ZNF706 ZNF706
## CORO1B.2 CORO1B
## TMEM59.2 TMEM59
## POLR1D.2 POLR1D
## RBM8A.1 RBM8A
## ATP5MC1 ATP5MC1
## TMED9.1 TMED9
## BLOC1S1.1 BLOC1S1
## EIF4A1 EIF4A1
## EIF3M.1 EIF3M
## MRPS24 MRPS24
## ANP32A.1 ANP32A
## ZNHIT1.2 ZNHIT1
## VDAC2 VDAC2
## MRPL41.1 MRPL41
## LAMTOR2.1 LAMTOR2
## CCT7 CCT7
## LIME1.2 LIME1
## ECH1.2 ECH1
## BSG.2 BSG
## TOMM5 TOMM5
## TRAPPC6A.2 TRAPPC6A
## REEP5.2 REEP5
## AP2S1.1 AP2S1
## VIM.2 VIM
## C11orf58.1 C11orf58
## GTF2A2.1 GTF2A2
## GIMAP7.2 GIMAP7
## NUTF2 NUTF2
## APH1A.1 APH1A
## MRPS34 MRPS34
## POLR2E.2 POLR2E
## LSM4.1 LSM4
## S1PR4.2 S1PR4
## LAMTOR1.1 LAMTOR1
## RNF181.1 RNF181
## ATP5MPL ATP5MPL
## ATRAID.2 ATRAID
## SRI.2 SRI
## CALR.2 CALR
## MICOS13.2 MICOS13
## TMEM219.1 TMEM219
## PPP1R18.2 PPP1R18
## RNF213.1 RNF213
## CCT2 CCT2
## ALYREF.1 ALYREF
## PSMD4.1 PSMD4
## NDUFA6.1 NDUFA6
## PSMA4.1 PSMA4
## SNRPF SNRPF
## MRPL54.1 MRPL54
## SIVA1.1 SIVA1
## TPM3.1 TPM3
## PRKCA.1 PRKCA
## TMEM14B TMEM14B
## C12orf75.2 C12orf75
## CACYBP.1 CACYBP
## SELENOF.2 SELENOF
## PDCD6.1 PDCD6
## ELOC.2 ELOC
## FKBP8.2 FKBP8
## MRPS15.1 MRPS15
## VAMP2.2 VAMP2
## RAB5IF RAB5IF
## POLR2G.2 POLR2G
## JPT1.1 JPT1
## CHURC1.2 CHURC1
## TKT.1 TKT
## NDUFB6 NDUFB6
## SUMO1.1 SUMO1
## KDELR1.1 KDELR1
## AURKAIP1.1 AURKAIP1
## PAIP2.2 PAIP2
## CTDNEP1.2 CTDNEP1
## MANF.1 MANF
## NME1.1 NME1
## NDUFB7.1 NDUFB7
## MRPL20 MRPL20
## TRAPPC5.1 TRAPPC5
## EID1.2 EID1
## EIF3D.1 EIF3D
## MAGOH.2 MAGOH
## CAPG.1 CAPG
## PSMB2.1 PSMB2
## ARHGDIA.1 ARHGDIA
## HOPX.2 HOPX
## HNRNPK.2 HNRNPK
## PPP1R14B.2 PPP1R14B
## TMEM230.1 TMEM230
## SNRPE.1 SNRPE
## ITM2A.2 ITM2A
## ALDOA ALDOA
## SET SET
## BCL2.1 BCL2
## TMED2.1 TMED2
## PSMB4.2 PSMB4
## SMIM26.2 SMIM26
## OTUB1.2 OTUB1
## SKP1.2 SKP1
## ANXA5.2 ANXA5
## DNPH1 DNPH1
## PA2G4.1 PA2G4
## STOML2 STOML2
## LAGE3.1 LAGE3
## PTGES3 PTGES3
## ANXA11.2 ANXA11
## ARF1.2 ARF1
## MT-ND2.2 MT-ND2
## HSP90AB1 HSP90AB1
## ALOX5AP.2 ALOX5AP
## TMEM14C TMEM14C
## PSMC5.1 PSMC5
## POLD4.2 POLD4
## DHRS7.2 DHRS7
## PDCD5 PDCD5
## PSMG2 PSMG2
## MRPL34.1 MRPL34
## RANBP1.2 RANBP1
## ETHE1.1 ETHE1
## MT2A.1 MT2A
## IGBP1.2 IGBP1
## MRPL43 MRPL43
## ETFB.1 ETFB
## HNRNPA0 HNRNPA0
## LSM6.1 LSM6
## ARL6IP5.2 ARL6IP5
## RNPS1 RNPS1
## SCP2.1 SCP2
## RNF167.2 RNF167
## CIAO2A.1 CIAO2A
## MYDGF.1 MYDGF
## SRM SRM
## MRPL57 MRPL57
## CCT3.1 CCT3
## VDAC1.1 VDAC1
## REXO2.2 REXO2
## H2AFV H2AFV
## PRMT1 PRMT1
## GADD45GIP1.1 GADD45GIP1
## PKM.1 PKM
## RBIS.1 RBIS
## MIEN1.1 MIEN1
## HMGB1 HMGB1
## NDUFV2.1 NDUFV2
## RNF7.2 RNF7
## PLSCR3.1 PLSCR3
## RHOG.2 RHOG
## ZNRD1.1 ZNRD1
## MRPL14 MRPL14
## COPS9.2 COPS9
## ILF2.1 ILF2
## AP2M1.2 AP2M1
## AIP.2 AIP
## CHMP2A.1 CHMP2A
## ADH5.2 ADH5
## DUT.2 DUT
## GIMAP1.2 GIMAP1
## SNRPD1.1 SNRPD1
## CMTM3.1 CMTM3
## PHB PHB
## EIF6.1 EIF6
## APEX1 APEX1
## XRCC6.1 XRCC6
## SPCS2.1 SPCS2
## UBE2B.2 UBE2B
## TUBA4A TUBA4A
## ACP1.2 ACP1
## RWDD1.1 RWDD1
## SHMT2 SHMT2
## TMEM123.1 TMEM123
## DCXR.2 DCXR
## ORAI1.2 ORAI1
## CLPP CLPP
## YPEL3.2 YPEL3
## UBE2V1.2 UBE2V1
## PSMD13.1 PSMD13
## NDUFAF3.1 NDUFAF3
## IFI27L2.1 IFI27L2
## MAP2K2.2 MAP2K2
## PSMC4.1 PSMC4
## NAP1L1.1 NAP1L1
## CAP1.2 CAP1
## ISG20.1 ISG20
## MBNL1.1 MBNL1
## PUF60.2 PUF60
## METTL26 METTL26
## ANXA2.2 ANXA2
## MRPL11 MRPL11
## CTSD.1 CTSD
## C1orf43 C1orf43
## SSNA1.1 SSNA1
## PSMD9.1 PSMD9
## LIMD2.2 LIMD2
## NBDY.1 NBDY
## MTPN.2 MTPN
## EIF1.2 EIF1
## RAD23A RAD23A
## IMP3.1 IMP3
## PTP4A2.2 PTP4A2
## FIBP.1 FIBP
## NKG7.2 NKG7
## SNRPB2.2 SNRPB2
## VDAC3 VDAC3
## GTF3C6.1 GTF3C6
## CD74.1 CD74
## MPG.2 MPG
## BIN1.2 BIN1
## MAT2B.2 MAT2B
## NPC2.1 NPC2
## HADHA.1 HADHA
## TMBIM4.1 TMBIM4
## TXN2 TXN2
## PSMC3.1 PSMC3
## SDHD SDHD
## SUPT4H1.1 SUPT4H1
## NDUFS3.1 NDUFS3
## STMP1.1 STMP1
## SSB SSB
## IDH2.2 IDH2
## TMEM256.1 TMEM256
## CNPY2.1 CNPY2
## BUD31.1 BUD31
## RPS19BP1 RPS19BP1
## NDUFB5.2 NDUFB5
## FAM174C FAM174C
## LSM3.1 LSM3
## PIN1.1 PIN1
## HMGN3.1 HMGN3
## ZNF428.1 ZNF428
## CSK.1 CSK
## PRDX3.1 PRDX3
## MPC2 MPC2
## CTSC.2 CTSC
## PPP2R1A.1 PPP2R1A
## TRADD.1 TRADD
## LSM5.2 LSM5
## CALM2.1 CALM2
## CCT8 CCT8
## COX14 COX14
## NUDT5.1 NUDT5
## CCT4 CCT4
## EMC4.1 EMC4
## PYURF.1 PYURF
## RPN2.1 RPN2
## SYNGR2 SYNGR2
## DEK.2 DEK
## HLA-DPA1.2 HLA-DPA1
## SELENOK.1 SELENOK
## HSD17B10.1 HSD17B10
## SUMO3.1 SUMO3
## SCAND1.1 SCAND1
## TMEM179B.1 TMEM179B
## SELENOT.1 SELENOT
## TXNL1.1 TXNL1
## AL138963.4.1 AL138963.4
## RNF187.1 RNF187
## HINT2 HINT2
## NAXE NAXE
## GSTO1 GSTO1
## PSMA3.1 PSMA3
## ADRM1.1 ADRM1
## TRAPPC2L.1 TRAPPC2L
## POLR2K.1 POLR2K
## UBE2N.1 UBE2N
## PPP1R7.1 PPP1R7
## RPN1 RPN1
## ENY2 ENY2
## COPS6.1 COPS6
## SDF2L1.1 SDF2L1
## ADSL ADSL
## RGS19.2 RGS19
## RAB5C.1 RAB5C
## STUB1.2 STUB1
## OXA1L.1 OXA1L
## SNF8 SNF8
## R3HDM4.2 R3HDM4
## VKORC1.1 VKORC1
## RPA3.2 RPA3
## FLT3LG.1 FLT3LG
## XBP1.2 XBP1
## MMADHC.1 MMADHC
## ABHD14B.2 ABHD14B
## DNAJC8.1 DNAJC8
## NDUFS6.1 NDUFS6
## SPCS3.1 SPCS3
## COA3.1 COA3
## AUP1.2 AUP1
## MLF2.1 MLF2
## TMEM35B.2 TMEM35B
## EIF2S3.2 EIF2S3
## KXD1 KXD1
## PTRHD1.1 PTRHD1
## ACAA2.1 ACAA2
## NONO NONO
## NENF.1 NENF
## MAZ.1 MAZ
## ARL6IP1.1 ARL6IP1
## GSTK1.1 GSTK1
## LSP1.2 LSP1
## SHISA5.2 SHISA5
## MT-ND6.2 MT-ND6
## SERPINB1.2 SERPINB1
## SF3B2.1 SF3B2
## SH3BGRL.2 SH3BGRL
## CD8B.2 CD8B
## CHMP4A.1 CHMP4A
## PDCD2 PDCD2
## NUCB2.1 NUCB2
## DSTN.2 DSTN
## DPY30.1 DPY30
## TCEA1.1 TCEA1
## LSM8.1 LSM8
## ZYX.2 ZYX
## MRPL52 MRPL52
## UQCRC1.1 UQCRC1
## SUCLG1 SUCLG1
## SEC13.1 SEC13
## PDIA3.1 PDIA3
## MEA1.1 MEA1
## PSMF1.1 PSMF1
## REX1BD.2 REX1BD
## NOL7 NOL7
## MRPS12 MRPS12
## PDIA6.1 PDIA6
## AKR1B1.1 AKR1B1
## UBE2D2.1 UBE2D2
## ARHGAP15 ARHGAP15
## WASHC3.2 WASHC3
## CCT5 CCT5
## SCAMP2.2 SCAMP2
## DCTN2.1 DCTN2
## PYCR2 PYCR2
## CLNS1A CLNS1A
## COX16.1 COX16
## TMEM238 TMEM238
## CCNDBP1.2 CCNDBP1
## SNX17.1 SNX17
## NSMCE1.1 NSMCE1
## CYC1 CYC1
## FAAP20.1 FAAP20
## LINC02694.2 LINC02694
## LAT.1 LAT
## CXCR3.2 CXCR3
## PABPN1.1 PABPN1
## PSMD7.1 PSMD7
## SLC9A3R1.2 SLC9A3R1
## PPIH.1 PPIH
## HLA-F.2 HLA-F
## TIMM13 TIMM13
## FDFT1.1 FDFT1
## URM1.1 URM1
## HAX1 HAX1
## EIF4B.1 EIF4B
## IAH1.1 IAH1
## KDELR2 KDELR2
## SARNP.1 SARNP
## TMEM147 TMEM147
## ILK.1 ILK
## SELPLG.2 SELPLG
## TEX264.1 TEX264
## MRPL4 MRPL4
## PTGER2.2 PTGER2
## EMC6.1 EMC6
## ARPC5L.1 ARPC5L
## BRI3 BRI3
## CLTB.1 CLTB
## CNPY3.1 CNPY3
## UBALD2.2 UBALD2
## CISD3.2 CISD3
## ZMAT2.1 ZMAT2
## CBX3.1 CBX3
## TMEM160.1 TMEM160
## GDI2 GDI2
## RAB1B.2 RAB1B
## RAB11B.2 RAB11B
## SERBP1.1 SERBP1
## COPZ1.1 COPZ1
## UBL7.1 UBL7
## CKLF.2 CKLF
## VPS29 VPS29
## TUBA1B.2 TUBA1B
## MRPS7 MRPS7
## PDE3B.2 PDE3B
## RER1.1 RER1
## EMD.1 EMD
## HSPB1.1 HSPB1
## GET3.2 GET3
## HPRT1.1 HPRT1
## TMBIM6.1 TMBIM6
## PCBP2 PCBP2
## SSU72.2 SSU72
## BZW1.2 BZW1
## CDKN2D.2 CDKN2D
## CARHSP1.1 CARHSP1
## CHCHD5.2 CHCHD5
## HNRNPA3 HNRNPA3
## ICAM2.2 ICAM2
## MRPS36.1 MRPS36
## THOC7 THOC7
## ABHD17A.2 ABHD17A
## FCMR.2 FCMR
## MFNG.2 MFNG
## RHOC.2 RHOC
## PSMB7 PSMB7
## CCDC167.2 CCDC167
## PFDN2 PFDN2
## C14orf119.1 C14orf119
## TNFSF12.1 TNFSF12
## AKR7A2.1 AKR7A2
## MRPL18 MRPL18
## NAPA.1 NAPA
## ANP32E.1 ANP32E
## SRP19.2 SRP19
## HLA-DPB1.2 HLA-DPB1
## CCNG1.1 CCNG1
## PCMT1.1 PCMT1
## IMPDH2.2 IMPDH2
## HMGB2.1 HMGB2
## CHCHD1.1 CHCHD1
## ROMO1 ROMO1
## FDPS.1 FDPS
## P4HB.1 P4HB
## SLC25A39 SLC25A39
## RBM42.1 RBM42
## PDAP1 PDAP1
## SNRPA SNRPA
## HSP90B1.1 HSP90B1
## MRPL21 MRPL21
## VSIR.1 VSIR
## TRAM1.2 TRAM1
## TXNDC17.2 TXNDC17
## GLRX.1 GLRX
## DOK2.2 DOK2
## EMC7.1 EMC7
## BUB3.1 BUB3
## TAP1.1 TAP1
## NUDT21.1 NUDT21
## DGCR6L DGCR6L
## CNIH1 CNIH1
## HMGA1.2 HMGA1
## LEPROTL1.1 LEPROTL1
## SMARCE1 SMARCE1
## STRAP.2 STRAP
## H1FX.1 H1FX
## MRPL16 MRPL16
## DGUOK.2 DGUOK
## PRKCH.1 PRKCH
## AQP3.2 AQP3
## TMED10.1 TMED10
## TRAC.2 TRAC
## ISG15.1 ISG15
## MRPL17.1 MRPL17
## SRSF5.2 SRSF5
## NUCKS1.2 NUCKS1
## PPM1G PPM1G
## THYN1.1 THYN1
## RHOF.1 RHOF
## HSBP1.1 HSBP1
## COMMD3.1 COMMD3
## COMMD7.1 COMMD7
## POLR2F.1 POLR2F
## NEAT1.1 NEAT1
## HSPB11.2 HSPB11
## MOSPD3.1 MOSPD3
## LAMP1.1 LAMP1
## TRBC1.2 TRBC1
## RPA2 RPA2
## RPL22L1 RPL22L1
## NUDCD2 NUDCD2
## LGALS3.2 LGALS3
## DMAC1 DMAC1
## METTL9.1 METTL9
## TMCO1.1 TMCO1
## CD47.2 CD47
## ECHS1.1 ECHS1
## CMPK1 CMPK1
## SARAF.1 SARAF
## CDC26.1 CDC26
## GLRX5 GLRX5
## ESD.1 ESD
## MESD MESD
## NUCB1.1 NUCB1
## WAS.2 WAS
## NDUFA8 NDUFA8
## CCDC107.1 CCDC107
## UQCRC2 UQCRC2
## ANKRD44 ANKRD44
## LYPLA2.1 LYPLA2
## NDUFS2.2 NDUFS2
## TMEM208 TMEM208
## CDC42.2 CDC42
## SDHAF2.1 SDHAF2
## NECAP2.2 NECAP2
## MRPS6.1 MRPS6
## CANX.1 CANX
## UFD1 UFD1
## CYB5R3.1 CYB5R3
## IER3IP1.2 IER3IP1
## PPP1CC PPP1CC
## NCR3.2 NCR3
## LSM10 LSM10
## RNF5.1 RNF5
## AKR1A1.1 AKR1A1
## SYF2.1 SYF2
## SF3A2.1 SF3A2
## SF3A1.1 SF3A1
## TMUB1 TMUB1
## CCDC85B.2 CCDC85B
## EIF1AY EIF1AY
## MRPS18C.1 MRPS18C
## SS18L2.1 SS18L2
## SEPTIN1.1 SEPTIN1
## MRPL13.1 MRPL13
## RNASET2 RNASET2
## COA4 COA4
## CNIH4 CNIH4
## PSMD6 PSMD6
## CUEDC2 CUEDC2
## BCL7C.2 BCL7C
## TSR3.1 TSR3
## H2AFY.1 H2AFY
## SLC25A11.2 SLC25A11
## TXNIP.2 TXNIP
## RALY RALY
## C12orf10 C12orf10
## JOSD2.1 JOSD2
## MRPS33.1 MRPS33
## IK.1 IK
## GCHFR.2 GCHFR
## HNRNPR HNRNPR
## EGLN2.2 EGLN2
## HNRNPD.1 HNRNPD
## PNKD.1 PNKD
## C9orf78.1 C9orf78
## MRFAP1L1.1 MRFAP1L1
## NAA38.1 NAA38
## ITPA.1 ITPA
## MRPS16.1 MRPS16
## ELP5 ELP5
## RBMX RBMX
## CASP4.2 CASP4
## HSPD1.2 HSPD1
## CTSW.2 CTSW
## UBXN4.1 UBXN4
## TMEM109.1 TMEM109
## HIKESHI HIKESHI
## PSENEN.1 PSENEN
## SNHG16.1 SNHG16
## UBE2M.1 UBE2M
## NDUFA3.1 NDUFA3
## CNPPD1.2 CNPPD1
## GOLGA7.1 GOLGA7
## NDUFV1.1 NDUFV1
## OCIAD1.1 OCIAD1
## SLC2A4RG SLC2A4RG
## MRPL33.1 MRPL33
## SH3BP1.2 SH3BP1
## TIMM8B.1 TIMM8B
## IFI6.1 IFI6
## OSTF1.2 OSTF1
## M6PR.1 M6PR
## CLTA CLTA
## MCTS1 MCTS1
## FUNDC2 FUNDC2
## THAP11 THAP11
## MEAF6.1 MEAF6
## PRPF19 PRPF19
## ZDHHC12 ZDHHC12
## DBNL.1 DBNL
## TAF9 TAF9
## CENPX.1 CENPX
## EVI2B.2 EVI2B
## NDUFB1 NDUFB1
## ARL2BP.1 ARL2BP
## TIMM17B TIMM17B
## HLA-E.2 HLA-E
## IDH3G.1 IDH3G
## PSIP1 PSIP1
## ZC3H15.2 ZC3H15
## CHMP5.1 CHMP5
## NUDC.1 NUDC
## PSMB8-AS1.2 PSMB8-AS1
## LYAR.2 LYAR
## CTSA.1 CTSA
## STX10 STX10
## MRPL36 MRPL36
## FKBP11.2 FKBP11
## VASP.1 VASP
## TMEM9B.2 TMEM9B
## POLE4 POLE4
## DNAJC19.1 DNAJC19
## SUSD3.1 SUSD3
## SMIM19 SMIM19
## METTL5 METTL5
## MAF1.1 MAF1
## COMT.1 COMT
## PPP1R2 PPP1R2
## DNAJA2.1 DNAJA2
## LITAF.2 LITAF
## ARF6.2 ARF6
## HDAC1 HDAC1
## NDUFA5.1 NDUFA5
## TRAPPC3.1 TRAPPC3
## SNHG32.1 SNHG32
## NIFK NIFK
## CARD16.1 CARD16
## DENND2D.1 DENND2D
## NDUFC1 NDUFC1
## FAM162A FAM162A
## HLA-DRB1.2 HLA-DRB1
## AGTRAP.2 AGTRAP
## NAA20 NAA20
## DCTPP1.2 DCTPP1
## CCDC124 CCDC124
## ARL2.2 ARL2
## SNX6 SNX6
## COA6 COA6
## POLR2H.1 POLR2H
## NUBP2 NUBP2
## PQBP1.1 PQBP1
## CD320 CD320
## ATG3 ATG3
## BLOC1S2.1 BLOC1S2
## SDHB SDHB
## GLRX3 GLRX3
## ZNHIT3.1 ZNHIT3
## TXNDC12.1 TXNDC12
## PHF5A PHF5A
## RNH1.1 RNH1
## SDF4.2 SDF4
## MACF1.2 MACF1
## PRELID3B PRELID3B
## POLR2I POLR2I
## GBP2.1 GBP2
## AP1S1.1 AP1S1
## AP1M1 AP1M1
## AHSA1.1 AHSA1
## COPS5.1 COPS5
## SRP72 SRP72
## FERMT3.1 FERMT3
## PIM1.2 PIM1
## H2AFJ.1 H2AFJ
## SCAMP3 SCAMP3
## FLOT1.2 FLOT1
## CDK4.2 CDK4
## POP5 POP5
## NDUFA9 NDUFA9
## FLOT2.2 FLOT2
## PPT1 PPT1
## CISD1 CISD1
## TIMM10.1 TIMM10
## ATP6V0D1.1 ATP6V0D1
## GRHPR.1 GRHPR
## MRPL28.1 MRPL28
## RPAIN RPAIN
## PTDSS1.1 PTDSS1
## FBXO7.1 FBXO7
## RCN2.1 RCN2
## BOLA3.1 BOLA3
## JUN.1 JUN
## SPAG7.1 SPAG7
## TADA3.1 TADA3
## ORMDL2.1 ORMDL2
## COX17 COX17
## SRA1 SRA1
## EIF1B.1 EIF1B
## PFDN6 PFDN6
## ELOF1.1 ELOF1
## SMIM12.1 SMIM12
## PSMC2.1 PSMC2
## ETFA.1 ETFA
## IGFLR1.1 IGFLR1
## MRPL9 MRPL9
## YWHAH.1 YWHAH
## PAFAH1B3.2 PAFAH1B3
## POLR3GL.1 POLR3GL
## HMGN4.2 HMGN4
## MOB1A.1 MOB1A
## ANXA7.1 ANXA7
## KLF2.1 KLF2
## TMEM126A TMEM126A
## MRPS35 MRPS35
## PFDN4 PFDN4
## VEGFB.2 VEGFB
## SELL.1 SELL
## SASH3.1 SASH3
## SNRPA1.1 SNRPA1
## NUDT1.2 NUDT1
## ECI1.1 ECI1
## FKBP3 FKBP3
## YIF1A.1 YIF1A
## LINC01871.2 LINC01871
## GPS2.2 GPS2
## CWC15 CWC15
## ATP6V0E2 ATP6V0E2
## CMC2 CMC2
## SRSF3 SRSF3
## UBE2A.1 UBE2A
## METTL23.2 METTL23
## LSM1.1 LSM1
## DNAJC7 DNAJC7
## COPS3.1 COPS3
## UFM1.1 UFM1
## PNRC1.2 PNRC1
## HNRNPF HNRNPF
## MIF4GD.1 MIF4GD
## HCFC1R1.1 HCFC1R1
## LYPLA1 LYPLA1
## SURF1.1 SURF1
## MRPL24 MRPL24
## HSP90AA1.1 HSP90AA1
## TSC22D4.1 TSC22D4
## WBP2.1 WBP2
## NDUFAF8.1 NDUFAF8
## PCNA.2 PCNA
## GRSF1.1 GRSF1
## UQCC2.2 UQCC2
## MRPL38.1 MRPL38
## RAB8A RAB8A
## RANGRF RANGRF
## DERL2.1 DERL2
## DNAJC15.1 DNAJC15
## GPS1 GPS1
## EIF2S2 EIF2S2
## EIF4E2.1 EIF4E2
## C18orf32.1 C18orf32
## BABAM1.1 BABAM1
## FAM32A FAM32A
## PLIN2.1 PLIN2
## RTF2.1 RTF2
## JAGN1 JAGN1
## CD8A.2 CD8A
## CRELD2.2 CRELD2
## ANXA6.2 ANXA6
## CTSZ CTSZ
## HTATIP2 HTATIP2
## ITGB7.2 ITGB7
## VBP1 VBP1
## ATP5ME ATP5ME
## DAXX.2 DAXX
## DRG1 DRG1
## NUDT22.1 NUDT22
## E2F4 E2F4
## TPRKB TPRKB
## C4orf48.1 C4orf48
## BAG1 BAG1
## GLO1.2 GLO1
## CDKN2A.2 CDKN2A
## MRPS18B MRPS18B
## MTCH2 MTCH2
## CSRP1.1 CSRP1
## IFI35.2 IFI35
## RPL26L1 RPL26L1
## CTSB.2 CTSB
## MPST.2 MPST
## RRP36 RRP36
## PPP6C PPP6C
## AC243960.1.2 AC243960.1
## HDDC2 HDDC2
## HTATSF1.1 HTATSF1
## SLBP.2 SLBP
## NELFE NELFE
## HDGF.1 HDGF
## RNASEH2B RNASEH2B
## EBPL EBPL
## CDC123.1 CDC123
## PRPF6 PRPF6
## MTLN MTLN
## LMAN1 LMAN1
## POP7.2 POP7
## FRG1 FRG1
## RBBP7 RBBP7
## ATP6AP2.1 ATP6AP2
## HNRNPM HNRNPM
## MAD2L2.2 MAD2L2
## ITGB1BP1.1 ITGB1BP1
## CAPZA2.1 CAPZA2
## MRPL12.1 MRPL12
## CAPZB.2 CAPZB
## HCLS1.2 HCLS1
## GYG1.1 GYG1
## DHRS4.1 DHRS4
## TPGS1.2 TPGS1
## COMMD8.1 COMMD8
## ZCRB1 ZCRB1
## STN1.1 STN1
## YIPF3.1 YIPF3
## TNFAIP8L2.1 TNFAIP8L2
## POLR3K.1 POLR3K
## PLEKHJ1.1 PLEKHJ1
## DOCK10.1 DOCK10
## TBC1D10C.2 TBC1D10C
## HNRNPUL1.1 HNRNPUL1
## SARS.1 SARS
## EEF1E1 EEF1E1
## TSTA3 TSTA3
## AP3S1 AP3S1
## PSMD11.1 PSMD11
## PARP1 PARP1
## DCPS.1 DCPS
## SMIM29.1 SMIM29
## BID BID
## MPLKIP MPLKIP
## PEF1.1 PEF1
## PPCS.1 PPCS
## TNFRSF14.1 TNFRSF14
## MRPL47 MRPL47
## CBR1.2 CBR1
## UROD UROD
## CEBPB.1 CEBPB
## TUBB4B TUBB4B
## APOBEC3C.1 APOBEC3C
## VMA21 VMA21
## GIMAP2.1 GIMAP2
## COMMD4.1 COMMD4
## CNOT7.1 CNOT7
## EXOSC1 EXOSC1
## DDA1.1 DDA1
## MED10.2 MED10
## UBA2.1 UBA2
## CPNE3.1 CPNE3
## PMPCB PMPCB
## UROS UROS
## RBM17 RBM17
## ZNF511 ZNF511
## MRPS23 MRPS23
## ABCE1 ABCE1
## WBP1 WBP1
## HAUS1 HAUS1
## MED28.1 MED28
## MFF MFF
## CD96.1 CD96
## MLX.1 MLX
## ANAPC15.1 ANAPC15
## PSMB5.1 PSMB5
## MCRS1.1 MCRS1
## CCNQ CCNQ
## MRPL22 MRPL22
## USP1.1 USP1
## MRPL3 MRPL3
## CD5 CD5
## ZNF22 ZNF22
## TMEM18 TMEM18
## ADIPOR1 ADIPOR1
## TIPRL.1 TIPRL
## TSR2 TSR2
## TMEM204.2 TMEM204
## CSDE1.2 CSDE1
## MRPL40 MRPL40
## SHKBP1.1 SHKBP1
## PCNP.1 PCNP
## IKZF1 IKZF1
## ELOVL1.1 ELOVL1
## MPV17 MPV17
## TMX1.1 TMX1
## WDR61 WDR61
## LSM14A.1 LSM14A
## AAMP AAMP
## RTN3 RTN3
## HLA-DMA.1 HLA-DMA
## TMEM14A TMEM14A
## SH2D2A.1 SH2D2A
## DUSP23.1 DUSP23
## PLAC8.2 PLAC8
## TRAPPC4 TRAPPC4
## RFXANK RFXANK
## DDRGK1 DDRGK1
## TERF2IP.1 TERF2IP
## CYB561D2.2 CYB561D2
## SQOR.1 SQOR
## STXBP2.2 STXBP2
## BCAS2 BCAS2
## HM13 HM13
## SSRP1.2 SSRP1
## HLA-DRA.2 HLA-DRA
## HDAC2.1 HDAC2
## ADI1.2 ADI1
## TCP1.1 TCP1
## AHCY.2 AHCY
## POP4.1 POP4
## MRPL58 MRPL58
## THOC3.1 THOC3
## POLR2C POLR2C
## TFDP1.2 TFDP1
## GBP4.1 GBP4
## SF3A3 SF3A3
## HACD4.1 HACD4
## COMTD1 COMTD1
## PNP.1 PNP
## SCNM1 SCNM1
## TRIAP1.1 TRIAP1
## VAPA.1 VAPA
## EBP EBP
## TMEM183A TMEM183A
## PPID PPID
## RUNX1.1 RUNX1
## YIF1B.1 YIF1B
## THEMIS.1 THEMIS
## CAMLG CAMLG
## IMP4 IMP4
## SSR1 SSR1
## TMEM134 TMEM134
## AIF1.1 AIF1
## EXOSC8.1 EXOSC8
## MRPS11 MRPS11
## RGL4.2 RGL4
## RAD21 RAD21
## TFPT.1 TFPT
## GMPR2 GMPR2
## SURF4.1 SURF4
## OXLD1 OXLD1
## MZT1.1 MZT1
## POLDIP2 POLDIP2
## PGD.2 PGD
## PRDX4.2 PRDX4
## VPS72.1 VPS72
## SNAPIN SNAPIN
## TINF2.1 TINF2
## TMEM126B TMEM126B
## ZFAND2B.1 ZFAND2B
## CELF2 CELF2
## ATIC.1 ATIC
## RELA.1 RELA
## AK2.1 AK2
## NMI NMI
## MFSD10.2 MFSD10
## ITGB2.2 ITGB2
## EIF2S1 EIF2S1
## EIF2A EIF2A
## SIRPG.1 SIRPG
## PTPN18 PTPN18
## MRPS2 MRPS2
## CDV3.1 CDV3
## BCCIP BCCIP
## VPS51.2 VPS51
## LYSMD2 LYSMD2
## CCDC59 CCDC59
## SDAD1 SDAD1
## GNAS.1 GNAS
## SKAP1.2 SKAP1
## TMEM106C.2 TMEM106C
## WDR54 WDR54
## MRPL27.1 MRPL27
## KPNB1.1 KPNB1
## SFT2D1.1 SFT2D1
## DPM2.2 DPM2
## MRPL15 MRPL15
## GIMAP5.2 GIMAP5
## CERS2 CERS2
## PSMD2 PSMD2
## CLDND1.2 CLDND1
## SNX5 SNX5
## RAB11A.1 RAB11A
## CTSS.2 CTSS
## ISCA2 ISCA2
## ASF1A ASF1A
## GLIPR2.1 GLIPR2
## MRFAP1 MRFAP1
## BECN1.1 BECN1
## MRPS26.1 MRPS26
## UPP1.2 UPP1
## AASDHPPT.1 AASDHPPT
## MRPL10.2 MRPL10
## DNAJB11 DNAJB11
## DPP7 DPP7
## BUD23 BUD23
## RNASEH2C.1 RNASEH2C
## ZNF524 ZNF524
## PDHB PDHB
## STIP1.2 STIP1
## FARSA FARSA
## CDIPT.1 CDIPT
## MRPL32 MRPL32
## FBXW5.2 FBXW5
## DOCK8.1 DOCK8
## TIMMDC1.1 TIMMDC1
## NUDT16L1.1 NUDT16L1
## HLA-DQB1.2 HLA-DQB1
## FNTA FNTA
## PPP1R11.1 PPP1R11
## NCL NCL
## PDZD11 PDZD11
## PPIE PPIE
## MGAT1.2 MGAT1
## NR1H2.1 NR1H2
## SNRNP40 SNRNP40
## LCK.2 LCK
## DDB1 DDB1
## DPYD DPYD
## ARL5A ARL5A
## MYCBP MYCBP
## MT-ATP8.2 MT-ATP8
## LAPTM4A.1 LAPTM4A
## EML4.2 EML4
## TMEM167A TMEM167A
## GMPS.1 GMPS
## ATP6AP1.1 ATP6AP1
## PSMD3.1 PSMD3
## TFAM TFAM
## RALA RALA
## C20orf27.2 C20orf27
## CCDC90B CCDC90B
## PMF1.1 PMF1
## PDCL3.1 PDCL3
## PCBD1.1 PCBD1
## SYNC SYNC
## RPP21 RPP21
## CYB5B CYB5B
## TCF7.2 TCF7
## BCKDHA BCKDHA
## CHST11.1 CHST11
## ISY1 ISY1
## SAMM50.1 SAMM50
## TIMM17A TIMM17A
## TRNAU1AP.1 TRNAU1AP
## UCHL5.1 UCHL5
## ELAVL1.1 ELAVL1
## DNAJC4.1 DNAJC4
## ABT1.1 ABT1
## PBDC1.1 PBDC1
## BLOC1S4.2 BLOC1S4
## BCL7B BCL7B
## AIMP1 AIMP1
## TMEM141.1 TMEM141
## STMN1.2 STMN1
## STT3A STT3A
## GNG10 GNG10
## MAD2L1.2 MAD2L1
## CISH.2 CISH
## TMEM173.2 TMEM173
## GTF2H5.1 GTF2H5
## SRGN.2 SRGN
## SNRNP25.2 SNRNP25
## MTFP1.1 MTFP1
## H2AFX.2 H2AFX
## PABPC1.2 PABPC1
## NCOA4.1 NCOA4
## GPAA1.2 GPAA1
## GRK6.1 GRK6
## SNRPN SNRPN
## KLHDC3 KLHDC3
## NDUFAF4 NDUFAF4
## UBE2V2 UBE2V2
## PRPF31 PRPF31
## HNRNPAB.2 HNRNPAB
## INO80E.1 INO80E
## PIGT.1 PIGT
## MTHFS.1 MTHFS
## PSMC1 PSMC1
## SLC25A1.1 SLC25A1
## MPC1 MPC1
## GLOD4 GLOD4
## BAD.1 BAD
## LCMT1 LCMT1
## LST1.2 LST1
## DHPS DHPS
## ATOX1.2 ATOX1
## MRPL37.1 MRPL37
## ZFAS1.2 ZFAS1
## PAGR1.1 PAGR1
## MAPKAPK5-AS1.1 MAPKAPK5-AS1
## MAPK1IP1L MAPK1IP1L
## CIRBP.2 CIRBP
## SRPRB SRPRB
## ACAT2.2 ACAT2
## CDK2AP1.2 CDK2AP1
## TMEM243.1 TMEM243
## CMTM7.1 CMTM7
## ZNRD2 ZNRD2
## ALDH9A1.1 ALDH9A1
## PTTG1.1 PTTG1
## MOB4.1 MOB4
## CAPN1.1 CAPN1
## DKC1 DKC1
## TXNDC9 TXNDC9
## C1orf122.1 C1orf122
## GTPBP6.1 GTPBP6
## MED30.1 MED30
## SEPTIN2.1 SEPTIN2
## RPL7L1 RPL7L1
## ANAPC13 ANAPC13
## RAB24.1 RAB24
## PNRC2 PNRC2
## SNHG7.1 SNHG7
## NTMT1 NTMT1
## YTHDF2 YTHDF2
## EIF3A.1 EIF3A
## MRPS14 MRPS14
## KARS KARS
## HMOX2.2 HMOX2
## RPS10 RPS10
## POLD2.2 POLD2
## GPR108.1 GPR108
## DTYMK.2 DTYMK
## CALCOCO2.2 CALCOCO2
## CYCS.1 CYCS
## ANKRD12 ANKRD12
## PSMC6 PSMC6
## VTI1B.2 VTI1B
## SIGIRR SIGIRR
## RFLNB.2 RFLNB
## CTBP1 CTBP1
## PSMG1.2 PSMG1
## CAPZA1.1 CAPZA1
## EBNA1BP2.1 EBNA1BP2
## MVP.1 MVP
## BTF3L4 BTF3L4
## SEPHS2.2 SEPHS2
## EIF1AX.1 EIF1AX
## HAGH.1 HAGH
## ZFAND1.1 ZFAND1
## PHF11 PHF11
## ABHD14A ABHD14A
## RBBP4 RBBP4
## BRMS1 BRMS1
## CDC25B.2 CDC25B
## PYM1 PYM1
## PDIA4 PDIA4
## ZNF581.1 ZNF581
## EAPP EAPP
## SIGMAR1 SIGMAR1
## UBC.2 UBC
## RUVBL1 RUVBL1
## CGGBP1 CGGBP1
## LAIR1.1 LAIR1
## COPS8 COPS8
## PAICS.2 PAICS
## PAPOLA PAPOLA
## GID8.1 GID8
## NSMCE3 NSMCE3
## PLD3.2 PLD3
## GSK3A.1 GSK3A
## NOB1 NOB1
## TSEN15.1 TSEN15
## PRPS1 PRPS1
## LGALS3BP.2 LGALS3BP
## NXT1.2 NXT1
## C1D C1D
## DECR1.1 DECR1
## UTRN.1 UTRN
## QARS QARS
## VPS25.1 VPS25
## MT-ND3.2 MT-ND3
## S1PR1.2 S1PR1
## NIT2.1 NIT2
## PRPF38A PRPF38A
## IDH3B IDH3B
## WDR77.2 WDR77
## USP39.1 USP39
## PKN1.1 PKN1
## CMAS CMAS
## NCF4.1 NCF4
## LRP10.2 LRP10
## ORMDL3.1 ORMDL3
## KRR1 KRR1
## ITM2C.2 ITM2C
## C15orf61 C15orf61
## MPDU1.1 MPDU1
## ANAPC5 ANAPC5
## PLGRKT PLGRKT
## HLA-DRB5.1 HLA-DRB5
## MITD1.1 MITD1
## AMZ2 AMZ2
## ACAA1 ACAA1
## G6PD.1 G6PD
## WDR18 WDR18
## SMARCD2 SMARCD2
## DCTD DCTD
## GGCT.2 GGCT
## DARS DARS
## MMP24OS.2 MMP24OS
## CCDC115.1 CCDC115
## RRM1.2 RRM1
## SNAP29 SNAP29
## MGMT MGMT
## MCRIP1.1 MCRIP1
## RAMAC RAMAC
## SLC35D2.2 SLC35D2
## HNRNPH2.1 HNRNPH2
## HDDC3 HDDC3
## KIF22.2 KIF22
## RAB18.2 RAB18
## B3GAT3 B3GAT3
## MAGED2.1 MAGED2
## TIMM50.1 TIMM50
## CYBC1.1 CYBC1
## NIPSNAP1 NIPSNAP1
## LTA4H LTA4H
## VPS35 VPS35
## MED11.1 MED11
## GTF2F1 GTF2F1
## LRRC59.1 LRRC59
## FAM50A.1 FAM50A
## PITHD1 PITHD1
## MID1IP1.1 MID1IP1
## TUSC2.1 TUSC2
## PPIG.2 PPIG
## APOBEC3H.2 APOBEC3H
## MRPL48 MRPL48
## RNF113A.1 RNF113A
## ZNF593.1 ZNF593
## ATXN1.1 ATXN1
## COMMD5.1 COMMD5
## CFAP298 CFAP298
## TSN TSN
## GOT2.1 GOT2
## PET100.1 PET100
## CD59 CD59
## FAM136A FAM136A
## CCDC28A.1 CCDC28A
## NTAN1.1 NTAN1
## PDE6D.1 PDE6D
## CCDC28B.1 CCDC28B
## MYC.2 MYC
## UBTF.1 UBTF
## STK16 STK16
## ANXA4.1 ANXA4
## NCBP2AS2.1 NCBP2AS2
## GNPDA1 GNPDA1
## SF1.1 SF1
## FDX1 FDX1
## NOP56 NOP56
## SLC3A2 SLC3A2
## FEN1.2 FEN1
## UBLCP1.1 UBLCP1
## BAK1 BAK1
## TMEM248.1 TMEM248
## ASAH1 ASAH1
## MALSU1.1 MALSU1
## SSBP4.2 SSBP4
## PFKL PFKL
## MCM7.2 MCM7
## SUPT16H.1 SUPT16H
## RAB7A.1 RAB7A
## MED8 MED8
## TIMM23.1 TIMM23
## NSFL1C NSFL1C
## LINC02446.2 LINC02446
## HSPA4 HSPA4
## TMEM223 TMEM223
## USB1 USB1
## CDC42SE2.1 CDC42SE2
## ACTR10.1 ACTR10
## RPS6KB2 RPS6KB2
## SUGT1 SUGT1
## DUS1L DUS1L
## CEP57 CEP57
## RPIA.1 RPIA
## DNAJC9.2 DNAJC9
## DAP3 DAP3
## EZR.2 EZR
## DDX39B.1 DDX39B
## COPB2 COPB2
## ATF4 ATF4
## TRAT1.1 TRAT1
## THOC6.1 THOC6
## OLA1.1 OLA1
## GALK1.1 GALK1
## SMC4.2 SMC4
## NOL12.1 NOL12
## HADH.2 HADH
## RILPL2 RILPL2
## FYTTD1 FYTTD1
## KEAP1.1 KEAP1
## UTP3 UTP3
## SMNDC1 SMNDC1
## CRBN.2 CRBN
## MRPL55 MRPL55
## SLC35B2 SLC35B2
## UBE2J2.1 UBE2J2
## MCM3.2 MCM3
## RPP25L RPP25L
## MOB2.2 MOB2
## DDIT4.2 DDIT4
## TXNDC15 TXNDC15
## TOR1A TOR1A
## AK6 AK6
## NGRN NGRN
## SPN.2 SPN
## CALHM2.1 CALHM2
## MTIF3.1 MTIF3
## CIAPIN1.1 CIAPIN1
## SMS.1 SMS
## GBP1.2 GBP1
## CZIB CZIB
## CCDC25 CCDC25
## MCRIP2 MCRIP2
## C7orf50.1 C7orf50
## ERG28 ERG28
## SVIP SVIP
## MBOAT7.2 MBOAT7
## COMMD9 COMMD9
## GLMP.1 GLMP
## MRPS10 MRPS10
## TOX4.1 TOX4
## COQ4 COQ4
## U2AF1L4 U2AF1L4
## ARL14EP.1 ARL14EP
## CS.1 CS
## RNPEPL1.2 RNPEPL1
## FTSJ1 FTSJ1
## RAE1 RAE1
## NOP16 NOP16
## INTS11 INTS11
## RWDD3 RWDD3
## MRPL2 MRPL2
## ALG3 ALG3
## ZNF24 ZNF24
## GNPTG GNPTG
## TIAL1 TIAL1
## ADD3.2 ADD3
## CD55.1 CD55
## GOLT1B GOLT1B
## C11orf1 C11orf1
## PAIP1 PAIP1
## SNAPC5 SNAPC5
## EI24 EI24
## PPM1M.1 PPM1M
## C16orf54.2 C16orf54
## RAB4A RAB4A
## SPTSSA SPTSSA
## RTCB.1 RTCB
## FBXL15 FBXL15
## EIF4EBP1.2 EIF4EBP1
## SKA2 SKA2
## EXOSC5.1 EXOSC5
## DNAAF2 DNAAF2
## SLC10A3.1 SLC10A3
## TRAF3IP3.2 TRAF3IP3
## CAT CAT
## MRPS25.2 MRPS25
## FH.2 FH
## NDUFS4 NDUFS4
## HERPUD1.1 HERPUD1
## HSD17B11.2 HSD17B11
## RNF220.1 RNF220
## ITGAE ITGAE
## SH2D1A.1 SH2D1A
## UBE2T.2 UBE2T
## ABI3.1 ABI3
## ADAM19.1 ADAM19
## DNASE2.1 DNASE2
## SAC3D1.2 SAC3D1
## TIMM9.1 TIMM9
## EIF2B1 EIF2B1
## HP1BP3.1 HP1BP3
## AKIP1.1 AKIP1
## TMEM205 TMEM205
## AKIRIN1.1 AKIRIN1
## DNPEP.1 DNPEP
## SRSF10.2 SRSF10
## ALG5 ALG5
## BCAT2.1 BCAT2
## RAB14.1 RAB14
## NABP2.1 NABP2
## GLTP.1 GLTP
## CDKN2AIPNL CDKN2AIPNL
## PDHA1 PDHA1
## SYNE2 SYNE2
## RRP7A.1 RRP7A
## PABPC4.2 PABPC4
## DIMT1 DIMT1
## COPS2.1 COPS2
## COA8 COA8
## SELENOS SELENOS
## ARL6IP6.1 ARL6IP6
## ISOC2.1 ISOC2
## SNX20.1 SNX20
## IMMT.1 IMMT
## DCK DCK
## ACTR3.1 ACTR3
## MRPL50 MRPL50
## UQCC3 UQCC3
## RPUSD3 RPUSD3
## UBA1 UBA1
## RNF114 RNF114
## SYPL1.2 SYPL1
## ACD.1 ACD
## UBE2S.1 UBE2S
## FLYWCH2 FLYWCH2
## EVI2A.1 EVI2A
## SLC66A3.1 SLC66A3
## MRPL42 MRPL42
## NINJ1.1 NINJ1
## ARL8A ARL8A
## FKBP4.2 FKBP4
## BRIX1.2 BRIX1
## YWHAE.2 YWHAE
## CCR7.2 CCR7
## CDC34.1 CDC34
## DENR DENR
## CDC42SE1.2 CDC42SE1
## BTG3 BTG3
## SLU7.1 SLU7
## GRPEL1.1 GRPEL1
## NFU1 NFU1
## ARMCX6 ARMCX6
## UBQLN2.2 UBQLN2
## POLR2J.1 POLR2J
## OPTN.2 OPTN
## CBX1.2 CBX1
## CHCHD7 CHCHD7
## MTX1 MTX1
## CD2BP2.1 CD2BP2
## SAP30BP SAP30BP
## SGF29 SGF29
## ARMC10 ARMC10
## CD69 CD69
## TRMT10C.1 TRMT10C
## LLPH LLPH
## XRN2.1 XRN2
## TMED5.1 TMED5
## SHFL.1 SHFL
## EPB41L4A-AS1 EPB41L4A-AS1
## EXOSC9.1 EXOSC9
## FAM204A FAM204A
## PSMG3.1 PSMG3
## YARS.2 YARS
## YDJC YDJC
## CD79B CD79B
## MRPS31 MRPS31
## COX11 COX11
## BLVRA.2 BLVRA
## SRSF6.1 SRSF6
## DLD.1 DLD
## YEATS4.2 YEATS4
## GTPBP4 GTPBP4
## SMIM7.1 SMIM7
## ZMAT5 ZMAT5
## LAMTOR3 LAMTOR3
## PRMT2.2 PRMT2
## TGFB1.2 TGFB1
## MRPS17.1 MRPS17
## MRPS18A MRPS18A
## SAT2.1 SAT2
## IL27RA IL27RA
## DNTTIP2.1 DNTTIP2
## MRPL46.1 MRPL46
## CDCA7.2 CDCA7
## PLPBP PLPBP
## SLC52A2.1 SLC52A2
## ATF6B.1 ATF6B
## DRG2 DRG2
## CKS1B.2 CKS1B
## GEMIN7.1 GEMIN7
## AP1S2 AP1S2
## PHAX.1 PHAX
## SNRNP70 SNRNP70
## GAMT.2 GAMT
## DYNLT3.1 DYNLT3
## SDHA SDHA
## COX20 COX20
## RAD51C.1 RAD51C
## WARS WARS
## API5 API5
## MAGOHB MAGOHB
## SLC39A3 SLC39A3
## DDX39A.2 DDX39A
## COPS4 COPS4
## HTRA2 HTRA2
## HRAS.1 HRAS
## C1orf162.2 C1orf162
## RABL6 RABL6
## C19orf48.2 C19orf48
## TMEM250 TMEM250
## AIMP2 AIMP2
## CRTAP CRTAP
## NGDN NGDN
## EFHD2.2 EFHD2
## HNRNPUL2.1 HNRNPUL2
## TMEM70.1 TMEM70
## AGPAT2.2 AGPAT2
## ENOPH1 ENOPH1
## FPGS FPGS
## HSPA9.1 HSPA9
## PCLAF.2 PCLAF
## MCM5.2 MCM5
## ARRDC1.1 ARRDC1
## WDR45.2 WDR45
## RWDD4 RWDD4
## SURF2 SURF2
## TOMM40.1 TOMM40
## UBL4A UBL4A
## RING1.1 RING1
## LBR LBR
## RHNO1 RHNO1
## HAT1.2 HAT1
## GRN.1 GRN
## COA5 COA5
## HAUS4.2 HAUS4
## FAM177A1.1 FAM177A1
## GSS.1 GSS
## RBM38.1 RBM38
## TCEAL8 TCEAL8
## PMVK.1 PMVK
## PXMP2.2 PXMP2
## PTPRC.2 PTPRC
## CLPTM1L.1 CLPTM1L
## UNG.2 UNG
## EIF4EBP2.1 EIF4EBP2
## AKTIP.1 AKTIP
## DYNLT1.1 DYNLT1
## MTCH1 MTCH1
## MRTO4.2 MRTO4
## STARD7 STARD7
## NUDT2 NUDT2
## RGS14.1 RGS14
## CEBPZOS.1 CEBPZOS
## LYRM2 LYRM2
## C11orf68.1 C11orf68
## FN3KRP.2 FN3KRP
## ANTKMT.2 ANTKMT
## ATPAF1.1 ATPAF1
## CFAP36 CFAP36
## ARL1 ARL1
## CKAP2.1 CKAP2
## PRKAG1 PRKAG1
## CDK5 CDK5
## TESC.2 TESC
## HSF1 HSF1
## PTBP1 PTBP1
## SLIRP SLIRP
## LIX1L LIX1L
## NOA1 NOA1
## TMEM203 TMEM203
## ARF3.2 ARF3
## RAD23B RAD23B
## APIP APIP
## PLRG1 PLRG1
## OGFR.1 OGFR
## PSME3.1 PSME3
## RRAGA.1 RRAGA
## CHMP1A CHMP1A
## COMMD1 COMMD1
## SINHCAF SINHCAF
## NAE1.1 NAE1
## HDHD5.1 HDHD5
## PDCD10.1 PDCD10
## DCUN1D5 DCUN1D5
## EXOSC4.2 EXOSC4
## DANCR.1 DANCR
## MAD1L1.1 MAD1L1
## SEPTIN6 SEPTIN6
## PSMG4 PSMG4
## PAF1 PAF1
## PIM3.1 PIM3
## FAM120AOS FAM120AOS
## TYMS.2 TYMS
## MRPL35.1 MRPL35
## INPP4B.2 INPP4B
## THAP7.1 THAP7
## CDKN1B.2 CDKN1B
## DPM3.1 DPM3
## ZDHHC24.1 ZDHHC24
## C1GALT1C1.1 C1GALT1C1
## CHID1 CHID1
## SDR39U1 SDR39U1
## C15orf40 C15orf40
## PSAP.2 PSAP
## PARL PARL
## MYD88.1 MYD88
## SPSB3.1 SPSB3
## SNHG5 SNHG5
## STYXL1 STYXL1
## LAG3.2 LAG3
## RAB33A.1 RAB33A
## NAPRT NAPRT
## SRSF8 SRSF8
## DNAJB1.1 DNAJB1
## PTGES2 PTGES2
## PPP6R1.1 PPP6R1
## CD40LG.2 CD40LG
## SH3GLB1.1 SH3GLB1
## TMEM60.1 TMEM60
## SAE1.2 SAE1
## TCEA2 TCEA2
## PARVG.1 PARVG
## CDIP1.1 CDIP1
## HEXB.1 HEXB
## CTDSP1.1 CTDSP1
## H3F3B.1 H3F3B
## CHTOP CHTOP
## TMX2.1 TMX2
## MRPL44.1 MRPL44
## NAGK NAGK
## PEPD.1 PEPD
## PIGP PIGP
## PRPF38B PRPF38B
## SERINC1.2 SERINC1
## AL133415.1 AL133415.1
## NIP7 NIP7
## SYTL1.2 SYTL1
## ID2.2 ID2
## MSN.1 MSN
## USP14.1 USP14
## CD164.1 CD164
## SDHAF3 SDHAF3
## GGNBP2 GGNBP2
## RAB2A RAB2A
## TP53TG1.1 TP53TG1
## SLC39A7 SLC39A7
## UBE2R2 UBE2R2
## TOR3A TOR3A
## IL16.2 IL16
## HMG20B HMG20B
## NANS.2 NANS
## EXOSC3.1 EXOSC3
## SERTAD1.1 SERTAD1
## LSM12 LSM12
## TMEM106B TMEM106B
## NPDC1.1 NPDC1
## YIPF5 YIPF5
## TMEM11.1 TMEM11
## RPA1 RPA1
## MTHFD2.2 MTHFD2
## MYL6B.1 MYL6B
## SLC39A1.1 SLC39A1
## RAB4B.1 RAB4B
## GMPPA GMPPA
## CASP1.1 CASP1
## MCUR1.1 MCUR1
## GUSB.1 GUSB
## SMIM15.1 SMIM15
## GMNN.2 GMNN
## MVB12A MVB12A
## AGGF1 AGGF1
## IFI30.1 IFI30
## FOPNL.1 FOPNL
## SNW1 SNW1
## PTMS.1 PTMS
## BORCS7 BORCS7
## TOMM70 TOMM70
## HIRIP3.2 HIRIP3
## TRABD.1 TRABD
## SCO2 SCO2
## FAM104A.1 FAM104A
## HACD3.1 HACD3
## BLVRB.1 BLVRB
## LMNB1.2 LMNB1
## CDC14A.1 CDC14A
## ENDOG ENDOG
## LARS LARS
## BTN3A2.2 BTN3A2
## RFC2.2 RFC2
## MT1X.1 MT1X
## ATP6V1D.1 ATP6V1D
## SMARCC2 SMARCC2
## ARFGAP3 ARFGAP3
## PEX16.1 PEX16
## IMPDH1 IMPDH1
## FAM200B FAM200B
## TNRC6B.1 TNRC6B
## VTA1 VTA1
## UTP11.1 UTP11
## GLS GLS
## TACC3.1 TACC3
## FTSJ3.1 FTSJ3
## DNAJC21.1 DNAJC21
## SNX15 SNX15
## SMC2.2 SMC2
## ACADM ACADM
## GLIPR1.1 GLIPR1
## HSPA1A.1 HSPA1A
## KRCC1 KRCC1
## IRF3 IRF3
## ACOT13 ACOT13
## MRPS5 MRPS5
## NKAP NKAP
## SEC61A1 SEC61A1
## SMIM3.2 SMIM3
## SLC25A20.1 SLC25A20
## ERCC1 ERCC1
## CAPRIN1.1 CAPRIN1
## AOAH.2 AOAH
## GPN1 GPN1
## BNIP2 BNIP2
## PIN4.2 PIN4
## DEDD2.1 DEDD2
## ELP6.1 ELP6
## NME4.2 NME4
## PTPMT1.1 PTPMT1
## CD151.1 CD151
## CYB5A.2 CYB5A
## GPKOW.1 GPKOW
## VRK1.2 VRK1
## TSFM.1 TSFM
## TMEM222 TMEM222
## COMMD2.1 COMMD2
## DHFR.2 DHFR
## TTC1 TTC1
## ZPR1 ZPR1
## PHF23.1 PHF23
## GINS2.2 GINS2
## CENPK.2 CENPK
## MRPL45.1 MRPL45
## GOLPH3.1 GOLPH3
## INPP5K.1 INPP5K
## TCTEX1D2.1 TCTEX1D2
## TOLLIP TOLLIP
## ACTL6A.1 ACTL6A
## SMPD1.1 SMPD1
## LZIC LZIC
## NPRL2.1 NPRL2
## TPP1.2 TPP1
## SMAD3.1 SMAD3
## MED4 MED4
## C5orf51 C5orf51
## AAAS.1 AAAS
## AATF AATF
## TBPL1 TBPL1
## NSMCE4A.1 NSMCE4A
## CINP CINP
## GFER GFER
## HSPBP1 HSPBP1
## EMC8 EMC8
## VAMP7.1 VAMP7
## MAP4K1.1 MAP4K1
## CDCA4.2 CDCA4
## SRRT.1 SRRT
## MRPL39 MRPL39
## QTRT1.1 QTRT1
## NUP37.2 NUP37
## SVBP SVBP
## RABGGTB RABGGTB
## RPP30.1 RPP30
## OVCA2.1 OVCA2
## ARFRP1 ARFRP1
## IKBIP.1 IKBIP
## TRIM69.2 TRIM69
## WDR46 WDR46
## AK3 AK3
## PGM1 PGM1
## GLRX2 GLRX2
## PAK2.1 PAK2
## PIGS.1 PIGS
## MTA2 MTA2
## MED16 MED16
## C19orf25.1 C19orf25
## POLR2D.1 POLR2D
## FBXO6.1 FBXO6
## COASY COASY
## IRF9 IRF9
## ZNF32 ZNF32
## NSUN5 NSUN5
## TPD52L2 TPD52L2
## IPO5.2 IPO5
## NBL1 NBL1
## ACOT8 ACOT8
## THUMPD3 THUMPD3
## AGPAT1 AGPAT1
## AP002387.2.1 AP002387.2
## TES.2 TES
## UBAC1 UBAC1
## BCL2L12.2 BCL2L12
## CDT1.2 CDT1
## TARS.1 TARS
## CCDC22 CCDC22
## ERAL1 ERAL1
## SOCS3.1 SOCS3
## POLR2B POLR2B
## RASSF1.2 RASSF1
## RBM22.1 RBM22
## IFRD2.1 IFRD2
## TMEM101 TMEM101
## KIF5B.2 KIF5B
## CUTC CUTC
## EFCAB14.1 EFCAB14
## GRB2 GRB2
## PEX11B PEX11B
## TMPO.2 TMPO
## BORCS6 BORCS6
## SCOC SCOC
## NELFCD.1 NELFCD
## CAMTA1 CAMTA1
## C22orf39 C22orf39
## NR2C2AP.2 NR2C2AP
## ACTR1A ACTR1A
## ANKRD39.1 ANKRD39
## EMG1 EMG1
## FASTK FASTK
## GIPC1 GIPC1
## NCF1 NCF1
## ATP6V1B2.2 ATP6V1B2
## TSEN54.1 TSEN54
## GPN3.2 GPN3
## EIF3B.1 EIF3B
## MTDH MTDH
## RABGAP1L RABGAP1L
## NRM.2 NRM
## TAF6.1 TAF6
## ARMC6 ARMC6
## LINC00667 LINC00667
## RBM4 RBM4
## CCDC57 CCDC57
## DNMT1.2 DNMT1
## LINC00623.2 LINC00623
## G3BP1 G3BP1
## EMB.2 EMB
## PPA2 PPA2
## ACYP1.1 ACYP1
## YBX3.2 YBX3
## MFSD1.1 MFSD1
## IKBKG IKBKG
## LTA LTA
## TXNDC5.1 TXNDC5
## BRCC3 BRCC3
## MPHOSPH10 MPHOSPH10
## EBAG9 EBAG9
## VPS26B VPS26B
## FIP1L1 FIP1L1
## CSNK1A1 CSNK1A1
## FAH.1 FAH
## FMC1 FMC1
## UPF3A.2 UPF3A
## YY1.1 YY1
## PEX2 PEX2
## PPIL3 PPIL3
## DEXI DEXI
## NRBF2 NRBF2
## SNHG1.1 SNHG1
## CHRAC1 CHRAC1
## DPH3 DPH3
## PLPP5 PLPP5
## GSKIP GSKIP
## SMIM20 SMIM20
## KLF6 KLF6
## GZMK.2 GZMK
## TMEM170A TMEM170A
## HOXB2.1 HOXB2
## SMC1A.1 SMC1A
## TAX1BP3.1 TAX1BP3
## RASSF7 RASSF7
## NOC2L NOC2L
## LRR1.2 LRR1
## TM2D1.1 TM2D1
## LRBA.1 LRBA
## NEPRO NEPRO
## CLK3 CLK3
## SYS1 SYS1
## CCNC CCNC
## ANXA1.2 ANXA1
## ZNF639 ZNF639
## MAD2L1BP MAD2L1BP
## CDC40 CDC40
## METAP2 METAP2
## NIPSNAP2.2 NIPSNAP2
## SMYD2 SMYD2
## MOCS2 MOCS2
## MCM4.2 MCM4
## SFXN1.1 SFXN1
## SGTA.1 SGTA
## APBA3.1 APBA3
## UBA3.1 UBA3
## SMARCA5 SMARCA5
## TOR2A.1 TOR2A
## ZWINT.2 ZWINT
## RGCC.1 RGCC
## METRN.1 METRN
## NUB1 NUB1
## GAR1 GAR1
## LINC02273.1 LINC02273
## DENND6B.1 DENND6B
## TAX1BP1.1 TAX1BP1
## LMF2.1 LMF2
## TRAP1.2 TRAP1
## EID2 EID2
## GDI1 GDI1
## STK25 STK25
## TRA2A TRA2A
## DR1 DR1
## WTAP WTAP
## CLEC2B.2 CLEC2B
## ARL4A ARL4A
## CHMP2B CHMP2B
## RBMX2 RBMX2
## ZSWIM7 ZSWIM7
## KAT8 KAT8
## CYB561A3 CYB561A3
## ERAP1 ERAP1
## GNAI3 GNAI3
## SLC35B1.1 SLC35B1
## ACAT1 ACAT1
## CCNB1IP1 CCNB1IP1
## EMC9.1 EMC9
## TSNAX TSNAX
## RASSF5.1 RASSF5
## ERVK3-1 ERVK3-1
## CLPTM1.1 CLPTM1
## DYRK4 DYRK4
## BRD7 BRD7
## SLC35A4 SLC35A4
## NIT1 NIT1
## TRIM8.1 TRIM8
## RP9 RP9
## EXOSC6 EXOSC6
## BTBD6.1 BTBD6
## SNAPC2 SNAPC2
## UBE2J1 UBE2J1
## EIF4E EIF4E
## ANKRD13A ANKRD13A
## PPIF.2 PPIF
## DOK1 DOK1
## SDHAF1 SDHAF1
## KIF2A.1 KIF2A
## MRM2 MRM2
## TYW3 TYW3
## OSGEP.1 OSGEP
## CASP6 CASP6
## MRPS22 MRPS22
## SNHG15 SNHG15
## FABP5.1 FABP5
## NIPSNAP3A NIPSNAP3A
## ZNF330 ZNF330
## IRF7.1 IRF7
## CDC5L CDC5L
## LDLRAP1.2 LDLRAP1
## EPS8L2.1 EPS8L2
## MRPS30 MRPS30
## ILF3 ILF3
## TMEM50B TMEM50B
## ARMC1 ARMC1
## WDR34.2 WDR34
## CHMP6.1 CHMP6
## COPB1 COPB1
## NTPCR NTPCR
## CXXC1.1 CXXC1
## PGRMC2 PGRMC2
## AGA AGA
## SNAP23 SNAP23
## RFC4.2 RFC4
## TMIGD2.1 TMIGD2
## GNPAT GNPAT
## ATP1B3.2 ATP1B3
## ARPP19.1 ARPP19
## SLC2A3 SLC2A3
## CCDC86.2 CCDC86
## MIB2.1 MIB2
## TESPA1.1 TESPA1
## CD70.2 CD70
## NIPA2 NIPA2
## ZNF622 ZNF622
## DCAF11.1 DCAF11
## UNC50 UNC50
## ARIH2 ARIH2
## NOP2 NOP2
## PHKG2.1 PHKG2
## RAP2B RAP2B
## AC008555.4.1 AC008555.4
## BAG6 BAG6
## GPATCH4 GPATCH4
## RNASEH2A.2 RNASEH2A
## RRM2.2 RRM2
## SP110.1 SP110
## RAB27A.1 RAB27A
## NUP107.2 NUP107
## PSMD10 PSMD10
## PCGF1 PCGF1
## DCAF7 DCAF7
## PDXP.1 PDXP
## GSPT1 GSPT1
## LAP3.1 LAP3
## SERPINB6.1 SERPINB6
## CLIC3.2 CLIC3
## EEF1AKMT2 EEF1AKMT2
## DEDD DEDD
## CITED2.2 CITED2
## GON7 GON7
## NKAPD1 NKAPD1
## BRCA1.2 BRCA1
## NUDT14.2 NUDT14
## ENO2.1 ENO2
## JMJD8 JMJD8
## SLC1A5.1 SLC1A5
## PIGX.1 PIGX
## MAP7D1.1 MAP7D1
## CAPN2.2 CAPN2
## CBR3.2 CBR3
## CDK1.2 CDK1
## C1orf131 C1orf131
## DNLZ DNLZ
## UNC45A.1 UNC45A
## BZW2 BZW2
## CPSF2 CPSF2
## MSH2.2 MSH2
## RSRP1.1 RSRP1
## CIAO1.1 CIAO1
## BEX4 BEX4
## PLAA PLAA
## NOC4L.1 NOC4L
## PRIM1.2 PRIM1
## DCAF13.1 DCAF13
## FBXO9.1 FBXO9
## CENPW.2 CENPW
## GART.1 GART
## PTTG1IP.1 PTTG1IP
## ACADS.1 ACADS
## ENSA ENSA
## GRAMD1A.1 GRAMD1A
## NMRAL1.1 NMRAL1
## NCOA2.1 NCOA2
## PRRC2A PRRC2A
## FAHD2B FAHD2B
## ESYT1.2 ESYT1
## DNTTIP1 DNTTIP1
## MRPL49 MRPL49
## OAS2 OAS2
## SLC4A1AP SLC4A1AP
## WRAP73 WRAP73
## SFPQ.1 SFPQ
## HIST3H2A.2 HIST3H2A
## ILKAP.1 ILKAP
## A1BG.1 A1BG
## ADPRHL2 ADPRHL2
## ASB8 ASB8
## PCIF1.1 PCIF1
## EBLN3P EBLN3P
## ZBTB7A.1 ZBTB7A
## LEPROT LEPROT
## RRS1.1 RRS1
## GTF2H3.1 GTF2H3
## PRKRA PRKRA
## TAPBP.1 TAPBP
## ERP44 ERP44
## SLC25A22 SLC25A22
## SCRN2 SCRN2
## CXorf40B.1 CXorf40B
## CHP1 CHP1
## SAP30 SAP30
## BDH2 BDH2
## NCAPD2.2 NCAPD2
## PSMA1 PSMA1
## TP53.2 TP53
## FAM98C.1 FAM98C
## ELK1 ELK1
## AARS.2 AARS
## PDRG1 PDRG1
## CARD19.1 CARD19
## KIF20B.2 KIF20B
## OARD1 OARD1
## CFAP20.1 CFAP20
## FAM110A FAM110A
## SEC22B.1 SEC22B
## HCP5 HCP5
## NSL1 NSL1
## CD82 CD82
## RAB21 RAB21
## ACADVL.1 ACADVL
## MRPL19 MRPL19
## LPCAT4.1 LPCAT4
## MSH6.2 MSH6
## COPRS.2 COPRS
## ZDHHC4 ZDHHC4
## ARFGAP2 ARFGAP2
## DMAC2 DMAC2
## UTP14A UTP14A
## CSNK1G2 CSNK1G2
## HIST1H1D.2 HIST1H1D
## C8orf76 C8orf76
## R3HCC1 R3HCC1
## TOR1AIP1.1 TOR1AIP1
## TM9SF1.1 TM9SF1
## RRP1B RRP1B
## PRCC PRCC
## ASF1B.2 ASF1B
## SART1 SART1
## XPA XPA
## TRPV2.2 TRPV2
## TMEM218.1 TMEM218
## NOP14.1 NOP14
## CENPH.2 CENPH
## CDC37 CDC37
## ERGIC2 ERGIC2
## DYNLL2 DYNLL2
## PIP4P1 PIP4P1
## TBRG1.1 TBRG1
## PIK3IP1.1 PIK3IP1
## CHMP3 CHMP3
## PARVB.1 PARVB
## DESI1.1 DESI1
## SMAP2.1 SMAP2
## DDX46 DDX46
## C12orf45.1 C12orf45
## THEM4.2 THEM4
## TK1.2 TK1
## PLSCR1 PLSCR1
## C12orf65 C12orf65
## RBM23.1 RBM23
## DRAM2 DRAM2
## SRPK1.1 SRPK1
## TAPBPL TAPBPL
## FLII FLII
## GFI1.1 GFI1
## TM2D2 TM2D2
## DDX42 DDX42
## HMCES HMCES
## LPGAT1 LPGAT1
## SWI5 SWI5
## ABTB1.2 ABTB1
## CNOT11 CNOT11
## JAML.1 JAML
## ALDH18A1 ALDH18A1
## MCM2.2 MCM2
## FBLN5 FBLN5
## RAB22A.1 RAB22A
## TRIM27 TRIM27
## TARBP2 TARBP2
## U2SURP U2SURP
## PRAF2 PRAF2
## TRUB2 TRUB2
## MPI.1 MPI
## EIF5B EIF5B
## SEPHS1.1 SEPHS1
## ABHD16A ABHD16A
## GMIP.1 GMIP
## CSE1L.2 CSE1L
## TCERG1.1 TCERG1
## MDP1 MDP1
## MAPK3.1 MAPK3
## KIAA2013.1 KIAA2013
## GOPC.1 GOPC
## IARS.2 IARS
## DDX23.1 DDX23
## RPUSD1.1 RPUSD1
## FAHD2A FAHD2A
## ITGAL.1 ITGAL
## TIMM29 TIMM29
## PGM2 PGM2
## TMF1.2 TMF1
## C16orf91 C16orf91
## RBBP8.2 RBBP8
## RETREG2.1 RETREG2
## DOHH DOHH
## LRPAP1.1 LRPAP1
## RPP38 RPP38
## RRAS RRAS
## ARL8B ARL8B
## LINC00892.2 LINC00892
## DNAJA3 DNAJA3
## ORAI3.1 ORAI3
## NELFB.1 NELFB
## INTS10 INTS10
## CIR1 CIR1
## YIPF2 YIPF2
## GPR65.1 GPR65
## MIA3 MIA3
## PTPRCAP.1 PTPRCAP
## PSMD14.1 PSMD14
## RABGGTA RABGGTA
## HPF1.1 HPF1
## BCAP29.1 BCAP29
## TRIM21 TRIM21
## MFSD5 MFSD5
## FAM3A FAM3A
## IRF2.1 IRF2
## CSTF1 CSTF1
## SMIM30 SMIM30
## BCKDK BCKDK
## RPF1 RPF1
## BATF.1 BATF
## PPHLN1 PPHLN1
## HSPA13 HSPA13
## ING4.1 ING4
## RTL8C.1 RTL8C
## RBBP6 RBBP6
## WDR82 WDR82
## P4HTM P4HTM
## PPIL1.2 PPIL1
## THAP1 THAP1
## ZCCHC9 ZCCHC9
## DTD1.1 DTD1
## ST6GALNAC4 ST6GALNAC4
## ZFAND2A ZFAND2A
## NOP58 NOP58
## UBN1 UBN1
## ISYNA1.2 ISYNA1
## PGRMC1.2 PGRMC1
## KYAT3.1 KYAT3
## PARP9.2 PARP9
## CD53.1 CD53
## NTHL1 NTHL1
## NEDD9.1 NEDD9
## FLAD1 FLAD1
## DGKZ.2 DGKZ
## MAP2K3 MAP2K3
## DMAC2L DMAC2L
## CTSH CTSH
## GET4.1 GET4
## NOL11.1 NOL11
## STMN3.1 STMN3
## GPR137 GPR137
## OCEL1 OCEL1
## STARD3NL.1 STARD3NL
## UNC119.1 UNC119
## DDX56.1 DDX56
## RALB RALB
## HSD17B8.1 HSD17B8
## GALT GALT
## PPP2R3C PPP2R3C
## MYH9.1 MYH9
## KATNBL1.1 KATNBL1
## CRLS1 CRLS1
## TPST2.2 TPST2
## DGAT1 DGAT1
## CNOT3 CNOT3
## NOLC1.2 NOLC1
## SRSF4 SRSF4
## PKMYT1.2 PKMYT1
## AC100810.1 AC100810.1
## ARAF.1 ARAF
## USE1 USE1
## ARMT1 ARMT1
## SEC23B.2 SEC23B
## CREB3 CREB3
## CD28.1 CD28
## HENMT1.1 HENMT1
## CDK9.1 CDK9
## USP16 USP16
## BSCL2 BSCL2
## YARS2 YARS2
## RFC1.1 RFC1
## TMUB2.1 TMUB2
## NFYC.1 NFYC
## SLC38A5 SLC38A5
## BNIP3L.1 BNIP3L
## SBDS.1 SBDS
## NAP1L4 NAP1L4
## PRKACA PRKACA
## EIF5 EIF5
## APOL3 APOL3
## DPH5.1 DPH5
## VPS36 VPS36
## MYADM.2 MYADM
## TXNRD1.1 TXNRD1
## CCDC12.1 CCDC12
## DPAGT1 DPAGT1
## MFAP1.1 MFAP1
## FGD5-AS1 FGD5-AS1
## TBCC.1 TBCC
## MOAP1 MOAP1
## CCL28 CCL28
## SNX1 SNX1
## HDGFL2.1 HDGFL2
## NBN.1 NBN
## ZNF580.1 ZNF580
## SUPT7L.1 SUPT7L
## SF3B1.1 SF3B1
## KAT2B.1 KAT2B
## ARFIP2.1 ARFIP2
## LYRM4.1 LYRM4
## CD300A.1 CD300A
## DNAJC2.2 DNAJC2
## PARP8.1 PARP8
## THOC2 THOC2
## NARS NARS
## TERF1.1 TERF1
## KLHDC2 KLHDC2
## ELOVL5 ELOVL5
## EIF4G1 EIF4G1
## SHC1 SHC1
## ABCF1 ABCF1
## ZFP36L1 ZFP36L1
## SAMHD1.2 SAMHD1
## KIAA1143 KIAA1143
## ASPSCR1 ASPSCR1
## PES1 PES1
## ARHGAP9.1 ARHGAP9
## C12orf76 C12orf76
## SNX10.1 SNX10
## SDCBP SDCBP
## PHGDH.2 PHGDH
## IDH3A.2 IDH3A
## ALG1 ALG1
## UHMK1 UHMK1
## FAM107B.1 FAM107B
## LIG1.2 LIG1
## RIOK3 RIOK3
## NCAPG2.2 NCAPG2
## PRMT5.1 PRMT5
## PITPNA.1 PITPNA
## CENPU.2 CENPU
## UBR7.1 UBR7
## PRPF40A.1 PRPF40A
## MRPL23.1 MRPL23
## ISCA1 ISCA1
## FOXN3 FOXN3
## KTN1 KTN1
## CHEK1.2 CHEK1
## TMEM199 TMEM199
## CIZ1 CIZ1
## BAZ1B.1 BAZ1B
## G3BP2.2 G3BP2
## WBP11.1 WBP11
## TGDS TGDS
## SLC44A2.1 SLC44A2
## DDX50.1 DDX50
## METTL21A.1 METTL21A
## PURA PURA
## FBXL16.1 FBXL16
## SAFB SAFB
## TRIM47 TRIM47
## ERI3 ERI3
## RAB9A.1 RAB9A
## DEAF1 DEAF1
## GLUD1 GLUD1
## BCL2A1.1 BCL2A1
## GPANK1 GPANK1
## GRWD1.1 GRWD1
## LHPP LHPP
## UHRF1.2 UHRF1
## HSPH1.2 HSPH1
## CGAS CGAS
## GPX7 GPX7
## TM9SF3.1 TM9SF3
## PRPSAP2 PRPSAP2
## BCAS4.2 BCAS4
## CRYL1 CRYL1
## IFT20 IFT20
## COQ5 COQ5
## GEMIN6 GEMIN6
## FAM111A.1 FAM111A
## CPSF4.1 CPSF4
## TSPAN3.1 TSPAN3
## ECSIT ECSIT
## OSTM1.1 OSTM1
## TEN1 TEN1
## GLT8D1 GLT8D1
## VHL VHL
## PIGH PIGH
## UTP6 UTP6
## LGALS9.2 LGALS9
## C1orf174 C1orf174
## CASP8AP2.1 CASP8AP2
## GSTM1.1 GSTM1
## RBM18.1 RBM18
## FAS.1 FAS
## MIIP.1 MIIP
## MYO1G.2 MYO1G
## DLST.1 DLST
## BCL11B BCL11B
## TMEM138 TMEM138
## CHTF8.1 CHTF8
## MARCH2.1 MARCH2
## CYSLTR1.2 CYSLTR1
## NPM3.2 NPM3
## STOM.2 STOM
## CHMP7 CHMP7
## DHX29 DHX29
## MAGED1 MAGED1
## LPXN.1 LPXN
## TUBA1A.1 TUBA1A
## SNRNP27 SNRNP27
## CSTF2T.1 CSTF2T
## KIAA1191.1 KIAA1191
## RBM7 RBM7
## SPOUT1 SPOUT1
## TNFSF14.2 TNFSF14
## IFT27.1 IFT27
## TPX2.2 TPX2
## SGO1.2 SGO1
## UTP18.1 UTP18
## PDK2 PDK2
## IFI16.1 IFI16
## ARV1 ARV1
## WSB2.1 WSB2
## SLC39A4.2 SLC39A4
## RNF214.1 RNF214
## EIF2D EIF2D
## GBP5.2 GBP5
## TGS1 TGS1
## TMEM128 TMEM128
## SNUPN.1 SNUPN
## FANCI.2 FANCI
## FAHD1 FAHD1
## ALAS1.1 ALAS1
## XAF1.1 XAF1
## MPHOSPH6.1 MPHOSPH6
## TRIM59 TRIM59
## BNIP3 BNIP3
## CCNK CCNK
## JMJD6 JMJD6
## NUP42.1 NUP42
## APEX2 APEX2
## RSRC2 RSRC2
## GCSH.1 GCSH
## DDX41 DDX41
## STARD3.1 STARD3
## VPS26A VPS26A
## ATG4B.1 ATG4B
## LMNB2.2 LMNB2
## PRKAB1 PRKAB1
## DUSP12 DUSP12
## OIP5-AS1 OIP5-AS1
## GTPBP8.1 GTPBP8
## RNPEP RNPEP
## SF3B3.1 SF3B3
## PPP5C.1 PPP5C
## GTF2H1 GTF2H1
## OSBP OSBP
## CCNA2.2 CCNA2
## GNL2.1 GNL2
## PELP1.1 PELP1
## TNFRSF25.1 TNFRSF25
## STK26.1 STK26
## C6orf89 C6orf89
## STAT5A.1 STAT5A
## MSRB2.2 MSRB2
## TIMM22 TIMM22
## CD4.2 CD4
## CEBPZ.1 CEBPZ
## MSRB1.1 MSRB1
## ACLY.1 ACLY
## SPINT2.2 SPINT2
## SLC27A5 SLC27A5
## LPP.1 LPP
## TUBA1C.1 TUBA1C
## ADAM15.1 ADAM15
## PARD6A PARD6A
## STK10.1 STK10
## TMEM189.1 TMEM189
## MYBL2.2 MYBL2
## BBC3.2 BBC3
## TBRG4 TBRG4
## LRIF1 LRIF1
## EEF1AKNMT EEF1AKNMT
## KPNA2.1 KPNA2
## HDLBP HDLBP
## ARMCX3 ARMCX3
## EXOC5 EXOC5
## GRINA GRINA
## C9orf40.2 C9orf40
## OTULINL.1 OTULINL
## TRABD2A.1 TRABD2A
## SGSM3 SGSM3
## RASA4 RASA4
## MUL1 MUL1
## NT5C3B.1 NT5C3B
## DDX27 DDX27
## SLC16A3.1 SLC16A3
## BARD1.2 BARD1
## OAT.2 OAT
## MED6.1 MED6
## ZMPSTE24 ZMPSTE24
## KLRK1.2 KLRK1
## UBE2C.2 UBE2C
## C6orf136 C6orf136
## COQ9 COQ9
## MGME1.1 MGME1
## ATP1A1 ATP1A1
## AC093323.1 AC093323.1
## RINL RINL
## FAM111B.2 FAM111B
## DUSP11.1 DUSP11
## DIAPH3.2 DIAPH3
## XAB2 XAB2
## AL441992.1 AL441992.1
## PGAP2.2 PGAP2
## RIOK2 RIOK2
## TPR TPR
## TRMT6 TRMT6
## SMIM27 SMIM27
## RRN3 RRN3
## JKAMP.1 JKAMP
## CTR9.1 CTR9
## CBX5.2 CBX5
## ZFP91.1 ZFP91
## CHRNB1 CHRNB1
## ALG8.2 ALG8
## TMEM259.2 TMEM259
## DHRS3.1 DHRS3
## ZMYND19.2 ZMYND19
## ZNF207 ZNF207
## DPP3.1 DPP3
## RDH14 RDH14
## MVD MVD
## LAPTM4B.1 LAPTM4B
## MLH1.1 MLH1
## UBA5 UBA5
## COPS7B COPS7B
## DAP.1 DAP
## OGFOD1 OGFOD1
## PDE7A.1 PDE7A
## KBTBD4 KBTBD4
## CHPF.1 CHPF
## PITPNA-AS1 PITPNA-AS1
## IFI27L1.2 IFI27L1
## C2orf68 C2orf68
## POLR3C POLR3C
## C2orf74 C2orf74
## RFX5 RFX5
## MAPK9.1 MAPK9
## ILVBL.1 ILVBL
## PRPF8 PRPF8
## LYRM1 LYRM1
## PPM1K PPM1K
## AIDA AIDA
## PAQR4.2 PAQR4
## PSMD1.1 PSMD1
## ACTR1B ACTR1B
## YBEY YBEY
## SNHG3 SNHG3
## SGO2.2 SGO2
## TMOD3.1 TMOD3
## ULK3 ULK3
## ISOC1.1 ISOC1
## CYREN CYREN
## TUBGCP2.1 TUBGCP2
## SBNO1.1 SBNO1
## HIST1H1B.2 HIST1H1B
## PIGBOS1.1 PIGBOS1
## C20orf204 C20orf204
## GLUL.1 GLUL
## TOP2A.2 TOP2A
## ASMTL ASMTL
## MT1F.1 MT1F
## UQCRHL UQCRHL
## RABEPK.1 RABEPK
## SLC43A3.2 SLC43A3
## TAF9B.1 TAF9B
## TDP2.1 TDP2
## CDC6.2 CDC6
## PRKCQ-AS1.2 PRKCQ-AS1
## POLR1E.1 POLR1E
## TBC1D10B.1 TBC1D10B
## AC022706.1.2 AC022706.1
## PDCL PDCL
## RRP9 RRP9
## DESI2.1 DESI2
## HEIH HEIH
## FAM117A.1 FAM117A
## DDAH2.1 DDAH2
## LRRC42.2 LRRC42
## TM9SF2.1 TM9SF2
## TACO1 TACO1
## TIFA.2 TIFA
## STAT1.2 STAT1
## GOLGA2 GOLGA2
## DDX6.1 DDX6
## PTPRJ.2 PTPRJ
## F11R F11R
## PARP10.1 PARP10
## DENND10 DENND10
## UCKL1 UCKL1
## UBIAD1 UBIAD1
## TP53RK TP53RK
## RTN4.1 RTN4
## ZNF326.1 ZNF326
## KRI1 KRI1
## MARCKSL1.1 MARCKSL1
## NFYB.1 NFYB
## CCDC47.1 CCDC47
## PLIN3.1 PLIN3
## ABLIM1.2 ABLIM1
## IL4R.1 IL4R
## CWC25 CWC25
## PDXK.1 PDXK
## MCFD2.1 MCFD2
## MMAB.2 MMAB
## B3GALT6 B3GALT6
## CGRRF1 CGRRF1
## PTPA PTPA
## SEC22C SEC22C
## SOCS2 SOCS2
## OAS1.1 OAS1
## TPRA1 TPRA1
## MAEA MAEA
## SAAL1.1 SAAL1
## TDG TDG
## DBP DBP
## ALG2 ALG2
## NCAPG.2 NCAPG
## MED19 MED19
## FAM217B FAM217B
## PTPN7.2 PTPN7
## ATL3.1 ATL3
## RUFY1 RUFY1
## TIMELESS.2 TIMELESS
## AHNAK.2 AHNAK
## MPP1.1 MPP1
## ZCCHC10 ZCCHC10
## IER5.1 IER5
## ANKRD11 ANKRD11
## ZNF410 ZNF410
## FARSB.1 FARSB
## CEBPG CEBPG
## PSPH PSPH
## MRS2.1 MRS2
## KATNA1 KATNA1
## GINM1 GINM1
## CDC45.2 CDC45
## UTP23.1 UTP23
## MED15.1 MED15
## ABCF3 ABCF3
## GLB1 GLB1
## GZMB.2 GZMB
## FAM210B FAM210B
## IGSF8.1 IGSF8
## KIFC1.2 KIFC1
## CNOT9.1 CNOT9
## DCTN1 DCTN1
## MYNN MYNN
## DMAP1 DMAP1
## KANSL1-AS1.1 KANSL1-AS1
## C8orf82 C8orf82
## MSMO1 MSMO1
## RCC1L.1 RCC1L
## VCP.1 VCP
## MORF4L2 MORF4L2
## ADA2.2 ADA2
## ARID1B.1 ARID1B
## MMGT1 MMGT1
## KIAA0040 KIAA0040
## PLEKHB2 PLEKHB2
## MRPS28.1 MRPS28
## KPNA3.1 KPNA3
## TSEN34 TSEN34
## SMARCC1.1 SMARCC1
## C8orf33.1 C8orf33
## TIMM10B TIMM10B
## BMS1 BMS1
## NMRK1 NMRK1
## AAGAB AAGAB
## CRTC2 CRTC2
## RIT1 RIT1
## TUBG1.2 TUBG1
## DPP4.1 DPP4
## UCK1.1 UCK1
## ORC6.2 ORC6
## LGALS8 LGALS8
## PDE6G PDE6G
## SLC7A6OS.1 SLC7A6OS
## YIPF4 YIPF4
## PPWD1 PPWD1
## THG1L THG1L
## MTMR14 MTMR14
## AARSD1 AARSD1
## PPP2R5D.1 PPP2R5D
## CCDC32 CCDC32
## NCAPD3.2 NCAPD3
## WDR55 WDR55
## PCYT2 PCYT2
## CTBS.1 CTBS
## SUDS3 SUDS3
## AZIN1.1 AZIN1
## CLSPN.2 CLSPN
## VRK3 VRK3
## CHST12.1 CHST12
## NCSTN NCSTN
## ODF2.2 ODF2
## POLDIP3 POLDIP3
## TSG101.1 TSG101
## SLC25A46 SLC25A46
## RAB1A RAB1A
## ZNF480 ZNF480
## KLHL9 KLHL9
## NELFA NELFA
## CHMP4B CHMP4B
## LPAR2 LPAR2
## TTC32 TTC32
## CTU1 CTU1
## TMEM80.1 TMEM80
## BBIP1 BBIP1
## DTX3L.1 DTX3L
## TNFRSF1B.1 TNFRSF1B
## G6PC3 G6PC3
## ATXN2L ATXN2L
## CTCF CTCF
## SHMT1.2 SHMT1
## ZNF830 ZNF830
## TBC1D17 TBC1D17
## ISG20L2 ISG20L2
## RHOT2 RHOT2
## TNFRSF1A.1 TNFRSF1A
## MED31 MED31
## TPMT.1 TPMT
## C1orf35.2 C1orf35
## C12orf43.1 C12orf43
## ACSL5 ACSL5
## TMEM216 TMEM216
## RRP1 RRP1
## PEMT.1 PEMT
## TRIB3 TRIB3
## FUOM.1 FUOM
## SRSF1.2 SRSF1
## LYPLAL1 LYPLAL1
## POLR1C.1 POLR1C
## TRAFD1 TRAFD1
## PPP1CB PPP1CB
## MRPL20-AS1 MRPL20-AS1
## SFXN4.2 SFXN4
## USP10 USP10
## LIN37.1 LIN37
## VPS33A VPS33A
## EIF3J EIF3J
## COA1 COA1
## GMCL1 GMCL1
## ZGPAT.1 ZGPAT
## POLQ.2 POLQ
## PFKP.2 PFKP
## TMEM167B TMEM167B
## SUPT20H.1 SUPT20H
## CCDC97 CCDC97
## BOP1 BOP1
## RPS6KA5.1 RPS6KA5
## CHKB CHKB
## RCSD1.1 RCSD1
## SRP54 SRP54
## CCDC58.1 CCDC58
## PDLIM1.1 PDLIM1
## WDR13 WDR13
## PHF6.1 PHF6
## FNDC3B.2 FNDC3B
## TM2D3 TM2D3
## CBFB.1 CBFB
## CPSF3.1 CPSF3
## SLC39A6.1 SLC39A6
## LTV1 LTV1
## CPTP CPTP
## ALG12 ALG12
## CITED4.1 CITED4
## CCAR1 CCAR1
## TIGIT.2 TIGIT
## TMX4.1 TMX4
## DHX9.1 DHX9
## LACTB2 LACTB2
## ALDOC ALDOC
## HAUS2.1 HAUS2
## C16orf87 C16orf87
## NOM1 NOM1
## GTF3C5.1 GTF3C5
## BLOC1S6 BLOC1S6
## RCE1 RCE1
## CLINT1 CLINT1
## CEACAM21.2 CEACAM21
## KNL1.2 KNL1
## ZDHHC3.1 ZDHHC3
## MAPRE2.2 MAPRE2
## TRAPPC2B TRAPPC2B
## PPAN.1 PPAN
## GMEB1 GMEB1
## UBE2E3.1 UBE2E3
## AP3D1 AP3D1
## BTBD9.1 BTBD9
## ZNF585A ZNF585A
## KCTD18 KCTD18
## PRPS2.2 PRPS2
## WDR36 WDR36
## AKIRIN2.1 AKIRIN2
## SH3YL1 SH3YL1
## ORAI2 ORAI2
## TWISTNB.1 TWISTNB
## SOD2.1 SOD2
## PRRC1 PRRC1
## GGH.2 GGH
## SYVN1 SYVN1
## CNOT8 CNOT8
## SLC35C2.1 SLC35C2
## AP5M1 AP5M1
## LRRC47.1 LRRC47
## BIN3.1 BIN3
## C6orf226.1 C6orf226
## ARL3.1 ARL3
## MIDN MIDN
## RUSC1.1 RUSC1
## EIF2B3.1 EIF2B3
## RIOK1 RIOK1
## MAGEH1 MAGEH1
## CTDSP2 CTDSP2
## CEP55.2 CEP55
## CRIPT CRIPT
## METTL17.1 METTL17
## CHPT1.1 CHPT1
## RPL39L.2 RPL39L
## BORCS8 BORCS8
## RDH11 RDH11
## RMC1 RMC1
## TOP1 TOP1
## SEC63 SEC63
## HSCB HSCB
## HCCS.1 HCCS
## TYSND1 TYSND1
## GSTM4 GSTM4
## TCIRG1.1 TCIRG1
## NAA80.1 NAA80
## UBE2D1.1 UBE2D1
## ATAD2.2 ATAD2
## ORC3.1 ORC3
## THEM6.1 THEM6
## IARS2 IARS2
## FANCG.2 FANCG
## MSI2.1 MSI2
## BET1 BET1
## PSAT1.2 PSAT1
## TMEM33 TMEM33
## SMCO4.1 SMCO4
## XPNPEP1.1 XPNPEP1
## ADORA2A ADORA2A
## PML PML
## DALRD3 DALRD3
## CCDC82 CCDC82
## FNDC10 FNDC10
## PCID2.1 PCID2
## KIF11.2 KIF11
## NUP88.1 NUP88
## IFNAR1 IFNAR1
## MND1.2 MND1
## WASHC1 WASHC1
## TMA16 TMA16
## NUP54.1 NUP54
## XCL2.2 XCL2
## NUBP1 NUBP1
## NMT1 NMT1
## MAP4K2 MAP4K2
## EWSR1 EWSR1
## SPNS1.1 SPNS1
## RFNG RFNG
## CENPS.2 CENPS
## MBD4 MBD4
## LSR.1 LSR
## PTS.1 PTS
## NFKBIZ.2 NFKBIZ
## KMT2C KMT2C
## ZDHHC2 ZDHHC2
## ZNF749 ZNF749
## WASHC5 WASHC5
## CARS.1 CARS
## TMEM115 TMEM115
## ATXN7L3B ATXN7L3B
## EFTUD2 EFTUD2
## PSTPIP1.1 PSTPIP1
## POC1A.2 POC1A
## TRA2B TRA2B
## BIRC5.2 BIRC5
## SIK3 SIK3
## TNFSF8.2 TNFSF8
## NELL2.1 NELL2
## MRRF MRRF
## RIDA.1 RIDA
## ACOT2 ACOT2
## ZNF598.1 ZNF598
## SIAH2 SIAH2
## TAZ TAZ
## ATG4D ATG4D
## DUS3L.1 DUS3L
## RAP1A.1 RAP1A
## REEP4.1 REEP4
## MRM3.1 MRM3
## PAM16 PAM16
## CYLD.1 CYLD
## ARHGAP30.2 ARHGAP30
## BAG5.2 BAG5
## CARS2.2 CARS2
## VAMP4.1 VAMP4
## THOC5 THOC5
## ATP8B4.2 ATP8B4
## PRADC1.1 PRADC1
## OGG1.1 OGG1
## MBIP MBIP
## PRPF4.1 PRPF4
## NUDT16.1 NUDT16
## DCAF12 DCAF12
## RIOX2 RIOX2
## TRIM11.1 TRIM11
## KCNA3.2 KCNA3
## FO393401.1.1 FO393401.1
## CDCA5.2 CDCA5
## TMEM42 TMEM42
## CDK5RAP3 CDK5RAP3
## VPS4B VPS4B
## SQSTM1.1 SQSTM1
## NT5DC1 NT5DC1
## DCTN6.1 DCTN6
## NUSAP1.2 NUSAP1
## PGS1 PGS1
## OGFOD2 OGFOD2
## DDX21.1 DDX21
## PCK2.1 PCK2
## BLCAP BLCAP
## REPIN1 REPIN1
## CENPV.1 CENPV
## TCF19.2 TCF19
## PLCB1.1 PLCB1
## SIKE1 SIKE1
## NRAS NRAS
## ZFAND3.1 ZFAND3
## GPATCH11.1 GPATCH11
## SUN2.2 SUN2
## PMM2 PMM2
## FAM102A.1 FAM102A
## TAF1D TAF1D
## DEF6.1 DEF6
## PMS2.1 PMS2
## ERCC5.1 ERCC5
## RCAN3AS RCAN3AS
## RRAGC RRAGC
## RAB5B.1 RAB5B
## FBXO45.1 FBXO45
## RAD1.2 RAD1
## GBA GBA
## TMEM99 TMEM99
## AC011603.2.2 AC011603.2
## CSTF2.1 CSTF2
## P2RX4.1 P2RX4
## VPS16 VPS16
## SLC39A8.2 SLC39A8
## ZC3H13.1 ZC3H13
## PTRH2.1 PTRH2
## CPSF7.1 CPSF7
## SLC66A2 SLC66A2
## NDUFAF2 NDUFAF2
## IFT52 IFT52
## TIA1 TIA1
## AC006064.4.2 AC006064.4
## OBI1.1 OBI1
## TMEM102.1 TMEM102
## PHF1.1 PHF1
## HSPA5 HSPA5
## TBP TBP
## DCAF15.1 DCAF15
## TMEM273.2 TMEM273
## PDF.2 PDF
## RANGAP1.2 RANGAP1
## TSC22D3.2 TSC22D3
## FAM89B FAM89B
## FNBP1 FNBP1
## STAT4.1 STAT4
## ZNF444 ZNF444
## TRIM22.2 TRIM22
## IPO7.2 IPO7
## STX17.1 STX17
## DHX36.1 DHX36
## LINC00299.2 LINC00299
## NIF3L1.1 NIF3L1
## CEP78.2 CEP78
## DNAJB12 DNAJB12
## IRF2BPL.1 IRF2BPL
## AP1B1.1 AP1B1
## CHMP1B CHMP1B
## MIER1.1 MIER1
## LRCH4 LRCH4
## GFM1 GFM1
## TBC1D4.1 TBC1D4
## STX4.1 STX4
## SCCPDH.2 SCCPDH
## SPOCK2.1 SPOCK2
## CAMK2G CAMK2G
## ETF1 ETF1
## KIF15.2 KIF15
## KCTD20.1 KCTD20
## C11orf54 C11orf54
## ZNF688 ZNF688
## IFNAR2 IFNAR2
## RBFA.2 RBFA
## PUSL1.1 PUSL1
## HAUS8.2 HAUS8
## CAMK1.1 CAMK1
## WDR5.2 WDR5
## TMEM120A TMEM120A
## AKT2 AKT2
## HPS6 HPS6
## XCL1.1 XCL1
## RNF34 RNF34
## CRLF3.1 CRLF3
## FBXL3 FBXL3
## DDX24 DDX24
## SYAP1 SYAP1
## CARD8 CARD8
## SLC35C1 SLC35C1
## MAP1S MAP1S
## CDKN3.2 CDKN3
## DPH2 DPH2
## LEO1.1 LEO1
## CLN3.1 CLN3
## CFP.1 CFP
## UBQLN4 UBQLN4
## ACY1 ACY1
## GUCD1.2 GUCD1
## GOSR2 GOSR2
## POMGNT1 POMGNT1
## ADCK2 ADCK2
## HBP1.1 HBP1
## PANK2 PANK2
## NKIRAS2 NKIRAS2
## SASS6.2 SASS6
## CASP2 CASP2
## TRAK2 TRAK2
## CEP63 CEP63
## COX19 COX19
## VPS37A VPS37A
## CDK2.2 CDK2
## SFT2D2 SFT2D2
## FBXO5.2 FBXO5
## PHYKPL.2 PHYKPL
## BTG1.2 BTG1
## FAF2 FAF2
## PCCB PCCB
## COQ6 COQ6
## CHAF1A.2 CHAF1A
## ARF4 ARF4
## CCDC92 CCDC92
## SUCLA2 SUCLA2
## PHLDA3.1 PHLDA3
## FUNDC1 FUNDC1
## GNPDA2 GNPDA2
## DPH1 DPH1
## WDR76.2 WDR76
## ADSS.1 ADSS
## TBC1D20 TBC1D20
## AC079793.1.1 AC079793.1
## TOPBP1.2 TOPBP1
## SIRT7 SIRT7
## MGAT2 MGAT2
## GTF2F2 GTF2F2
## FAM104B FAM104B
## EIF2B2 EIF2B2
## CDCA8.2 CDCA8
## DAPP1 DAPP1
## PXN.2 PXN
## MKNK2 MKNK2
## HNRNPU HNRNPU
## YTHDF1.1 YTHDF1
## EML2 EML2
## ZFAND6.1 ZFAND6
## KLRD1.2 KLRD1
## GBP3 GBP3
## L3MBTL2.1 L3MBTL2
## PPIL4 PPIL4
## SIPA1.1 SIPA1
## FOXM1.2 FOXM1
## NFKBIL1 NFKBIL1
## RAB6A RAB6A
## REEP3.2 REEP3
## RTL8A RTL8A
## AURKB.2 AURKB
## TMEM209.1 TMEM209
## TMEM69.1 TMEM69
## EHMT2.1 EHMT2
## BROX BROX
## MARS MARS
## RO60 RO60
## RAD51AP1.2 RAD51AP1
## STAU1 STAU1
## ARHGAP1 ARHGAP1
## PDCD6IP PDCD6IP
## CCDC137.1 CCDC137
## LIPA LIPA
## NDUFAF1.1 NDUFAF1
## RITA1.1 RITA1
## RNF10 RNF10
## PEA15.1 PEA15
## RAVER1.1 RAVER1
## SART3 SART3
## TAMM41 TAMM41
## ALKBH6 ALKBH6
## AMDHD2 AMDHD2
## PCGF6.2 PCGF6
## PAK1IP1.1 PAK1IP1
## HDHD2 HDHD2
## MCM10.2 MCM10
## C21orf91.2 C21orf91
## SNAI3 SNAI3
## LINC002481.2 LINC002481
## TRMT1 TRMT1
## PAG1.1 PAG1
## BTN3A3 BTN3A3
## DNAJB6 DNAJB6
## UBALD1 UBALD1
## CFDP1 CFDP1
## CIP2A.2 CIP2A
## COPS7A COPS7A
## PSMD5 PSMD5
## PCGF5.1 PCGF5
## CRYZ CRYZ
## P2RY10 P2RY10
## NAPG NAPG
## GTF2E2 GTF2E2
## AC026347.1.1 AC026347.1
## SH3GL1 SH3GL1
## RACGAP1.2 RACGAP1
## HYOU1 HYOU1
## C2orf49 C2orf49
## GNL1.1 GNL1
## PELO PELO
## UBE2Q2 UBE2Q2
## DUSP2.2 DUSP2
## MTFMT MTFMT
## TEDC1.2 TEDC1
## MFSD12.1 MFSD12
## RNF8.1 RNF8
## NEDD1.2 NEDD1
## KLF3.2 KLF3
## PRR14 PRR14
## GPALPP1 GPALPP1
## CARD8-AS1 CARD8-AS1
## ZCCHC8 ZCCHC8
## TRMT61B TRMT61B
## WAC-AS1.1 WAC-AS1
## HK1 HK1
## IDS.1 IDS
## TMEM251 TMEM251
## USP30-AS1 USP30-AS1
## PPP4R2 PPP4R2
## ATP13A1 ATP13A1
## AKT1S1 AKT1S1
## TP53I13 TP53I13
## MCMBP.1 MCMBP
## ZNF787 ZNF787
## ZDHHC16.1 ZDHHC16
## PXMP4 PXMP4
## CHCHD4 CHCHD4
## DHRS1.1 DHRS1
## GNB1 GNB1
## PDCD4.1 PDCD4
## RELB.1 RELB
## TSPAN17.1 TSPAN17
## JMJD7.1 JMJD7
## BYSL BYSL
## MTERF4 MTERF4
## UBAP2L.1 UBAP2L
## HSPA1B HSPA1B
## B3GNT2.1 B3GNT2
## OTUD5 OTUD5
## ZNF277.1 ZNF277
## ATAD5.2 ATAD5
## PRKAR1B.1 PRKAR1B
## RNF25 RNF25
## NAALADL1 NAALADL1
## CYTH2 CYTH2
## ZFPL1.1 ZFPL1
## ZDHHC18 ZDHHC18
## CBLB.1 CBLB
## TRIM38.1 TRIM38
## INTS14 INTS14
## EXOC4.1 EXOC4
## AC079781.5 AC079781.5
## C12orf49 C12orf49
## CWC27 CWC27
## RAB39B RAB39B
## CCDC43 CCDC43
## GNB5 GNB5
## DNAJC17.1 DNAJC17
## TATDN3 TATDN3
## EIF4G2.1 EIF4G2
## RAPGEF1.1 RAPGEF1
## DCP2 DCP2
## MCCC2 MCCC2
## CDCA2.2 CDCA2
## CYB5R1 CYB5R1
## LINC00909 LINC00909
## KRT7.1 KRT7
## MTG1 MTG1
## UBXN11.2 UBXN11
## AKAP13.1 AKAP13
## ANAPC7 ANAPC7
## ERCC3 ERCC3
## TMEM97.2 TMEM97
## PARPBP.2 PARPBP
## BRI3BP.2 BRI3BP
## UPF3B UPF3B
## EML3 EML3
## RABL3 RABL3
## FAM114A2 FAM114A2
## ZNF544.1 ZNF544
## SCAF1.1 SCAF1
## IMMP1L.1 IMMP1L
## TMEM254 TMEM254
## MARCH5 MARCH5
## GOT1.1 GOT1
## SEPTIN9.1 SEPTIN9
## EIF2AK2.1 EIF2AK2
## ARSA.1 ARSA
## HEBP1.2 HEBP1
## NCBP3.1 NCBP3
## EVA1B.1 EVA1B
## LINC01578 LINC01578
## CPB2-AS1.1 CPB2-AS1
## INTS5 INTS5
## SLC41A3 SLC41A3
## POLD1.2 POLD1
## ATAD1 ATAD1
## MBD2.1 MBD2
## SLA2.1 SLA2
## TIMM8A TIMM8A
## DNAL4 DNAL4
## NUP93.1 NUP93
## CTNNBIP1.1 CTNNBIP1
## NADK NADK
## SRSF2.1 SRSF2
## ZNF414 ZNF414
## TCP11L2.2 TCP11L2
## APOL2.1 APOL2
## RXRB RXRB
## LINC00339 LINC00339
## XRCC1.1 XRCC1
## ITPRIPL1.1 ITPRIPL1
## KIAA0825.1 KIAA0825
## EDEM2 EDEM2
## RMI2.2 RMI2
## DUSP1.2 DUSP1
## OIP5.2 OIP5
## SMC6.1 SMC6
## MPV17L2 MPV17L2
## ATP6V0A2 ATP6V0A2
## BIRC2 BIRC2
## IFT57 IFT57
## TMED7 TMED7
## ING3 ING3
## KIN KIN
## WBP4 WBP4
## WEE1.2 WEE1
## STX8.2 STX8
## MIS18A.2 MIS18A
## SLC29A1.2 SLC29A1
## TFAP4.2 TFAP4
## STYX STYX
## GCAT.1 GCAT
## KMT5A.2 KMT5A
## ME2.1 ME2
## TTC4 TTC4
## WAPL WAPL
## RNF138.1 RNF138
## IL10RB IL10RB
## PDLIM7.1 PDLIM7
## UBP1 UBP1
## CETN3.1 CETN3
## SLC30A5 SLC30A5
## MRPS9 MRPS9
## ARID5A ARID5A
## TMEM187 TMEM187
## C18orf21 C18orf21
## FUCA2 FUCA2
## LY96.1 LY96
## PLOD3 PLOD3
## CCNB1.2 CCNB1
## TTC28-AS1 TTC28-AS1
## HEMK1 HEMK1
## MANEA.2 MANEA
## PUS7L PUS7L
## RARS RARS
## EGLN3.1 EGLN3
## TTC37 TTC37
## ACBD3 ACBD3
## IRAK4.1 IRAK4
## SLC22A18.1 SLC22A18
## DGKA.2 DGKA
## PWWP2A PWWP2A
## INTS8 INTS8
## ZNF720 ZNF720
## BRCA2.2 BRCA2
## SNX18.1 SNX18
## NLE1 NLE1
## ZRANB2 ZRANB2
## DNAJC30 DNAJC30
## BRIP1.2 BRIP1
## ELAC2.1 ELAC2
## MAT2A MAT2A
## PNO1 PNO1
## ABRAXAS1 ABRAXAS1
## CSNK2A2 CSNK2A2
## HEXD HEXD
## CDC42EP3.2 CDC42EP3
## DYNC1LI1 DYNC1LI1
## AZI2 AZI2
## PRXL2C PRXL2C
## NPAT.1 NPAT
## MELK.2 MELK
## INO80C INO80C
## STAP1.1 STAP1
## YY1AP1 YY1AP1
## CCDC18-AS1 CCDC18-AS1
## LMBRD1 LMBRD1
## SNHG30 SNHG30
## SPC25.2 SPC25
## BLOC1S5 BLOC1S5
## ACSL3 ACSL3
## TBC1D10A TBC1D10A
## MED24.1 MED24
## SQLE.1 SQLE
## MVK.1 MVK
## EIF1AD EIF1AD
## UBE2E1.2 UBE2E1
## SPATS2L.1 SPATS2L
## CCDC61 CCDC61
## CDKN2AIP.1 CDKN2AIP
## GET1.1 GET1
## PAPSS1.1 PAPSS1
## ASCC2 ASCC2
## KDM1A.1 KDM1A
## ATP6V1H ATP6V1H
## NMD3 NMD3
## HMGB3.2 HMGB3
## MTAP.1 MTAP
## ICOS.1 ICOS
## BRD4.1 BRD4
## MPPE1 MPPE1
## EMC2.1 EMC2
## TCTN3 TCTN3
## IRAK1.1 IRAK1
## C1orf52 C1orf52
## MAPKAPK5 MAPKAPK5
## PEX19 PEX19
## DSN1.2 DSN1
## CTNNBL1 CTNNBL1
## RANBP2.1 RANBP2
## MT1E.1 MT1E
## AP1G2.1 AP1G2
## HAGHL.1 HAGHL
## NUP58.2 NUP58
## TMEM9.1 TMEM9
## MSL3 MSL3
## CHORDC1.1 CHORDC1
## TMEM8A TMEM8A
## TARDBP TARDBP
## FBH1 FBH1
## MED1 MED1
## MTIF2 MTIF2
## ZNF800 ZNF800
## NDUFV3 NDUFV3
## C19orf12.1 C19orf12
## SLC25A24 SLC25A24
## DIPK1A.1 DIPK1A
## MAGT1.1 MAGT1
## MECR.1 MECR
## MFGE8.1 MFGE8
## NUF2.2 NUF2
## FBRS.1 FBRS
## ZBTB24 ZBTB24
## PTPN11 PTPN11
## ALDH6A1 ALDH6A1
## AEN AEN
## KLRC4.2 KLRC4
## DLAT DLAT
## N4BP2L1 N4BP2L1
## ZNF76 ZNF76
## DYNC1I2.1 DYNC1I2
## FAM210A FAM210A
## ALAD ALAD
## ALKBH2.1 ALKBH2
## AP5S1 AP5S1
## TRIOBP.1 TRIOBP
## PREPL PREPL
## CASTOR1 CASTOR1
## RABEP1 RABEP1
## TBL3 TBL3
## ZNF384 ZNF384
## AL138724.1 AL138724.1
## RTF1 RTF1
## ZNF101.1 ZNF101
## CMTM8.2 CMTM8
## RNF168.2 RNF168
## MIS18BP1 MIS18BP1
## DPEP2.1 DPEP2
## SRXN1 SRXN1
## RAB5A.1 RAB5A
## PRR5.2 PRR5
## GTF3C2.1 GTF3C2
## CCDC71L.1 CCDC71L
## NAGPA NAGPA
## SLK.1 SLK
## TADA1 TADA1
## OXCT1.2 OXCT1
## ANKRD54 ANKRD54
## METAP1.1 METAP1
## LINC00861.2 LINC00861
## HJURP.2 HJURP
## COQ2.1 COQ2
## URI1.1 URI1
## OOEP OOEP
## DHX15.1 DHX15
## TELO2.2 TELO2
## DERA DERA
## UBXN2A.1 UBXN2A
## SLC33A1 SLC33A1
## ATP23.1 ATP23
## SLC25A33 SLC25A33
## KIF2C.2 KIF2C
## CAND1.1 CAND1
## CDK10 CDK10
## EIF4EBP3.1 EIF4EBP3
## KLRB1.2 KLRB1
## PHTF1 PHTF1
## RAD50 RAD50
## GANAB GANAB
## ESCO1 ESCO1
## BCLAF1 BCLAF1
## SEC23IP SEC23IP
## EXOC3 EXOC3
## MPND.1 MPND
## ZC3H7A.1 ZC3H7A
## NAA50 NAA50
## RNF11.1 RNF11
## MUS81 MUS81
## STK17B.1 STK17B
## ST6GALNAC6.1 ST6GALNAC6
## RAMP1.1 RAMP1
## MKI67.2 MKI67
## SFMBT2.2 SFMBT2
## ASPM.2 ASPM
## ARHGAP19.2 ARHGAP19
## GEMIN4.2 GEMIN4
## CCDC127 CCDC127
## MTRES1.1 MTRES1
## LRRC14 LRRC14
## NEIL3.2 NEIL3
## ANKRD13D ANKRD13D
## CYTOR.1 CYTOR
## ETFDH ETFDH
## SMIM4 SMIM4
## LDOC1 LDOC1
## AC012146.1 AC012146.1
## GALNT1.1 GALNT1
## INCENP.2 INCENP
## C17orf75.1 C17orf75
## WSB1.1 WSB1
## POLR2M POLR2M
## IBTK IBTK
## DTWD1 DTWD1
## HSD17B4.1 HSD17B4
## YIPF6 YIPF6
## SNAP47.1 SNAP47
## CYP20A1.1 CYP20A1
## SLFN12L SLFN12L
## SLC25A32 SLC25A32
## CD46.2 CD46
## YIPF1.1 YIPF1
## MAN1B1.1 MAN1B1
## ANLN.2 ANLN
## BLM.2 BLM
## BRD3.1 BRD3
## QRICH1 QRICH1
## DTNBP1 DTNBP1
## STAMBP STAMBP
## OSER1.1 OSER1
## ZC3H8 ZC3H8
## SPRYD3.1 SPRYD3
## TMEM214 TMEM214
## C15orf39 C15orf39
## CENPL.2 CENPL
## DNAJB14.1 DNAJB14
## RCN1 RCN1
## PPP4R3A.1 PPP4R3A
## FAM122B FAM122B
## SAMD9.2 SAMD9
## NT5C3A NT5C3A
## CCDC34.2 CCDC34
## HDGFL3 HDGFL3
## TMEM263 TMEM263
## REXO4 REXO4
## CDK5RAP1 CDK5RAP1
## BTBD1 BTBD1
## AC013264.1 AC013264.1
## NR2F6.1 NR2F6
## RRP8 RRP8
## POLB POLB
## TIPIN.2 TIPIN
## DDX52 DDX52
## CD38.2 CD38
## ARFIP1 ARFIP1
## ABCD4 ABCD4
## WDR48 WDR48
## ANKRA2 ANKRA2
## DYRK2.1 DYRK2
## AP3M1 AP3M1
## STIL.2 STIL
## CASP3 CASP3
## WDR12.1 WDR12
## LDB1 LDB1
## TRIP13.2 TRIP13
## PERP.2 PERP
## NOS3.1 NOS3
## NUDT8.1 NUDT8
## LINC01003 LINC01003
## OMA1 OMA1
## UBE2G2 UBE2G2
## CKAP2L.2 CKAP2L
## BUB1.2 BUB1
## GATA3 GATA3
## SLC25A23 SLC25A23
## HIC1.2 HIC1
## PGAM5.1 PGAM5
## BEX3.1 BEX3
## AP2A1.1 AP2A1
## FMNL1.2 FMNL1
## PITPNB PITPNB
## ABRAXAS2 ABRAXAS2
## JMJD4.2 JMJD4
## IST1 IST1
## PHC2 PHC2
## C7orf26 C7orf26
## PNPT1 PNPT1
## CHAC2.2 CHAC2
## LIAS LIAS
## MYBBP1A.1 MYBBP1A
## AC010642.2 AC010642.2
## TMEM165 TMEM165
## CAAP1 CAAP1
## NCLN.1 NCLN
## CWF19L1 CWF19L1
## RAD51.2 RAD51
## PPP1R3F.1 PPP1R3F
## USP8 USP8
## CDC7.2 CDC7
## ARMH1.2 ARMH1
## GIMAP8.1 GIMAP8
## RXYLT1.1 RXYLT1
## RRP15 RRP15
## RAB10 RAB10
## GABPB1-IT1 GABPB1-IT1
## AP5Z1 AP5Z1
## ADPGK ADPGK
## MAP11 MAP11
## TUT1.1 TUT1
## CENPE.2 CENPE
## CEP41.1 CEP41
## BNIP1.1 BNIP1
## PTCD3.1 PTCD3
## UBE2Z UBE2Z
## PREX1.1 PREX1
## NAT10 NAT10
## XPC.2 XPC
## GTF2H4 GTF2H4
## PHF20L1 PHF20L1
## ELP2 ELP2
## SRFBP1 SRFBP1
## GIT1 GIT1
## SRD5A3.1 SRD5A3
## MTF2 MTF2
## RNF4.1 RNF4
## SCAMP4 SCAMP4
## TOPORS TOPORS
## ZC3H14 ZC3H14
## IMPAD1 IMPAD1
## SLC25A36 SLC25A36
## MYL5 MYL5
## NAA60 NAA60
## IFIH1.1 IFIH1
## PLAAT3.2 PLAAT3
## SAMD1.2 SAMD1
## PNKP.1 PNKP
## TMEM154 TMEM154
## NQO2.1 NQO2
## COX10 COX10
## DCUN1D1.1 DCUN1D1
## SLAIN2.1 SLAIN2
## TMEM129 TMEM129
## EEF2KMT.1 EEF2KMT
## EXOSC10 EXOSC10
## IFT22.1 IFT22
## ZNF770 ZNF770
## SECISBP2L.1 SECISBP2L
## DENND1C.1 DENND1C
## IKBKE IKBKE
## CCNF.2 CCNF
## PDXDC1.1 PDXDC1
## RASSF2 RASSF2
## DHX30.1 DHX30
## DARS2 DARS2
## POGK POGK
## IL2RB.1 IL2RB
## TIGAR.1 TIGAR
## MMUT.1 MMUT
## CEP104 CEP104
## CIAO3 CIAO3
## MED25 MED25
## KAT2A.1 KAT2A
## LSG1.1 LSG1
## CDC23.1 CDC23
## HSBP1L1 HSBP1L1
## KCTD10 KCTD10
## CENPN.2 CENPN
## GOSR1 GOSR1
## RNF115 RNF115
## CLK4 CLK4
## ARL15.1 ARL15
## TRIM4 TRIM4
## HAUS6.1 HAUS6
## ING5.1 ING5
## GCFC2 GCFC2
## LAS1L.1 LAS1L
## ELMO1.2 ELMO1
## AC093157.1.2 AC093157.1
## TMEM159.1 TMEM159
## BEX2.1 BEX2
## IP6K2 IP6K2
## RFK RFK
## AAR2.1 AAR2
## CHD4 CHD4
## ALDH3A2 ALDH3A2
## DUSP28 DUSP28
## TRAF2.2 TRAF2
## ARRDC2 ARRDC2
## PMS1 PMS1
## CEP19.2 CEP19
## SURF6 SURF6
## JPT2.2 JPT2
## PDCD2L PDCD2L
## NXT2 NXT2
## GGA1.1 GGA1
## AC016831.7.1 AC016831.7
## SNX4.1 SNX4
## SAMD10.1 SAMD10
## HSH2D.1 HSH2D
## BPNT1 BPNT1
## NUDCD1 NUDCD1
## ZNF433-AS1 ZNF433-AS1
## MAIP1 MAIP1
## HLTF.1 HLTF
## MAP2K1.1 MAP2K1
## ZFYVE27 ZFYVE27
## NDC1.2 NDC1
## DHRS4-AS1 DHRS4-AS1
## GSTZ1.1 GSTZ1
## GLI4 GLI4
## TNKS2 TNKS2
## TSPAN32.2 TSPAN32
## TAF1B TAF1B
## DYNC1LI2 DYNC1LI2
## SMAD2 SMAD2
## CHAF1B.2 CHAF1B
## OAZ2.1 OAZ2
## ELK4 ELK4
## PARP3.1 PARP3
## TSR1.2 TSR1
## ZNF106 ZNF106
## SLTM SLTM
## SOCS1.1 SOCS1
## PRKAA1 PRKAA1
## PPP3CA.2 PPP3CA
## EXOC6 EXOC6
## KATNB1.1 KATNB1
## ZNF721 ZNF721
## ESRRA.1 ESRRA
## ZNF236-DT ZNF236-DT
## INTS12 INTS12
## PNPO PNPO
## SFT2D3 SFT2D3
## MBD3.2 MBD3
## MAP4K5 MAP4K5
## MGAT5.2 MGAT5
## NBEAL1 NBEAL1
## NOL8 NOL8
## C12orf29 C12orf29
## METTL18 METTL18
## TOB1.1 TOB1
## RRAGD.2 RRAGD
## FOXRED1.1 FOXRED1
## CDC27.1 CDC27
## CHCHD3.1 CHCHD3
## COG4 COG4
## UBE2D4 UBE2D4
## ITGB1.2 ITGB1
## ING1 ING1
## ZNF217.2 ZNF217
## GALE.1 GALE
## SCLY.1 SCLY
## SREK1IP1 SREK1IP1
## SMG6.1 SMG6
## HCFC1.2 HCFC1
## ASCC1 ASCC1
## TRGV10.2 TRGV10
## STARD10.1 STARD10
## ACOT9 ACOT9
## RAB35.1 RAB35
## RMDN3 RMDN3
## TMEM127 TMEM127
## LINC00847 LINC00847
## FAM98A FAM98A
## TBL2 TBL2
## USO1 USO1
## BTN3A1.1 BTN3A1
## TKFC.1 TKFC
## HGH1 HGH1
## LIPE LIPE
## ZMYM5 ZMYM5
## COPA.1 COPA
## TRIB2 TRIB2
## CCPG1.1 CCPG1
## C6orf47 C6orf47
## ZFR ZFR
## FANCD2.2 FANCD2
## RBM25 RBM25
## CDC20.2 CDC20
## UBE2G1.1 UBE2G1
## RBM14.1 RBM14
## FAM50B.1 FAM50B
## OAS3.2 OAS3
## CHERP CHERP
## IL18BP IL18BP
## LINC00324.2 LINC00324
## ALKBH3.1 ALKBH3
## CCR4.2 CCR4
## RMDN1 RMDN1
## CLN5.2 CLN5
## MTERF3.1 MTERF3
## MTFR1L.1 MTFR1L
## TRMT2A TRMT2A
## TXLNG TXLNG
## RNF20 RNF20
## SAP30L SAP30L
## CIT.2 CIT
## OLFM2 OLFM2
## CCDC18.2 CCDC18
## GTPBP3.1 GTPBP3
## USP22.2 USP22
## PIAS4 PIAS4
## CWC22.1 CWC22
## BX890604.2 BX890604.2
## ZNF880 ZNF880
## LIPT1 LIPT1
## TMC6.1 TMC6
## SH2B1 SH2B1
## LACTB.1 LACTB
## TRIM5 TRIM5
## SPAG16.2 SPAG16
## JOSD1.2 JOSD1
## NBR1 NBR1
## MAGEF1.1 MAGEF1
## XPOT.2 XPOT
## RAB3GAP1.2 RAB3GAP1
## ESCO2.2 ESCO2
## ECHDC2.2 ECHDC2
## ZFYVE21.1 ZFYVE21
## NIPAL3 NIPAL3
## LFNG.1 LFNG
## DDX1 DDX1
## RSBN1 RSBN1
## TMEM107.1 TMEM107
## SUV39H1.2 SUV39H1
## SPDL1.2 SPDL1
## SAFB2 SAFB2
## PXN-AS1.1 PXN-AS1
## HBS1L.1 HBS1L
## TTI2.1 TTI2
## UBXN6 UBXN6
## TMEM64.1 TMEM64
## DLGAP5.2 DLGAP5
## RPF2.1 RPF2
## GIT2.1 GIT2
## PINX1 PINX1
## ICMT.2 ICMT
## TRAF3IP2.1 TRAF3IP2
## RAD9A.1 RAD9A
## ZNF576.1 ZNF576
## UBE3A UBE3A
## GPR155.1 GPR155
## MCM8.2 MCM8
## LUC7L3 LUC7L3
## ESS2.1 ESS2
## ZNF766 ZNF766
## SKP2.2 SKP2
## USP48.1 USP48
## DIS3L.1 DIS3L
## SMPD4.1 SMPD4
## KCNK6 KCNK6
## TMED1 TMED1
## RCHY1 RCHY1
## AAMDC.1 AAMDC
## APPBP2 APPBP2
## CHST14 CHST14
## MEN1 MEN1
## DBR1 DBR1
## GOLGA5 GOLGA5
## CTNNAL1.2 CTNNAL1
## TMEM120B TMEM120B
## FAM133B FAM133B
## CENPB.1 CENPB
## ZFPM1 ZFPM1
## FAM219B.1 FAM219B
## HLA-DMB.1 HLA-DMB
## CMTR2 CMTR2
## LRRC41.1 LRRC41
## KLHDC4 KLHDC4
## KIF23.2 KIF23
## CREBL2 CREBL2
## USP28.1 USP28
## DBNDD2.2 DBNDD2
## GTSE1.2 GTSE1
## C11orf24.1 C11orf24
## LIMS1 LIMS1
## TMEM140 TMEM140
## SLC25A26 SLC25A26
## SPATC1L.1 SPATC1L
## CHAMP1.1 CHAMP1
## WHAMM WHAMM
## DDX20 DDX20
## EXOC7 EXOC7
## AC245297.3 AC245297.3
## SFR1.1 SFR1
## ZSCAN16-AS1 ZSCAN16-AS1
## HHLA3 HHLA3
## USF2.1 USF2
## INIP.2 INIP
## MKRN2 MKRN2
## RHOH RHOH
## CPSF1 CPSF1
## KPNA4 KPNA4
## SPOP SPOP
## KHSRP.2 KHSRP
## YWHAZ.2 YWHAZ
## TXK.1 TXK
## CDKN2C.2 CDKN2C
## RAP2A.1 RAP2A
## MAP3K13.1 MAP3K13
## PIGK PIGK
## PPP1R15A.1 PPP1R15A
## RAD54L.2 RAD54L
## CLPX CLPX
## SELENOM.2 SELENOM
## ZNHIT2 ZNHIT2
## CCDC71 CCDC71
## TTC3 TTC3
## NPRL3.1 NPRL3
## KRIT1 KRIT1
## SLC19A1.2 SLC19A1
## CASC3 CASC3
## NPLOC4.1 NPLOC4
## FCER1G FCER1G
## WRNIP1 WRNIP1
## SHPRH.1 SHPRH
## RNF141 RNF141
## FASTKD2 FASTKD2
## TVP23B TVP23B
## LETM1 LETM1
## CKAP5.1 CKAP5
## REST.1 REST
## KHK.2 KHK
## RBBP5.1 RBBP5
## KLHL22.1 KLHL22
## KIAA1586 KIAA1586
## TRIM52-AS1 TRIM52-AS1
## ORC5 ORC5
## RWDD2B RWDD2B
## TMEM87B TMEM87B
## TSPYL1 TSPYL1
## TRMT61A TRMT61A
## SNX14.1 SNX14
## PCED1A.1 PCED1A
## DCAF8 DCAF8
## ATAD3A.2 ATAD3A
## EEA1 EEA1
## CDCA7L.2 CDCA7L
## FANCB.2 FANCB
## PRKACB.1 PRKACB
## CFD CFD
## MTRF1L MTRF1L
## COIL COIL
## POGLUT1 POGLUT1
## SUPV3L1.1 SUPV3L1
## QRSL1.1 QRSL1
## PRC1.2 PRC1
## PECAM1.2 PECAM1
## SKA3.2 SKA3
## IFT43.1 IFT43
## CREM.1 CREM
## SPAG5.2 SPAG5
## AP003774.2 AP003774.2
## TBC1D7 TBC1D7
## CASP7 CASP7
## ZMYND11 ZMYND11
## TSPAN14.1 TSPAN14
## CYP51A1 CYP51A1
## SELENOO SELENOO
## SFXN3 SFXN3
## GLE1 GLE1
## SPART.1 SPART
## HIF1AN HIF1AN
## SEPTIN8.1 SEPTIN8
## CD2AP.2 CD2AP
## ACAD8 ACAD8
## GCH1.1 GCH1
## ZBED5 ZBED5
## LASP1.1 LASP1
## CCDC186.1 CCDC186
## ASB1 ASB1
## BAG3.2 BAG3
## GTF2I.1 GTF2I
## EIF2AK4 EIF2AK4
## POLRMT POLRMT
## BAG4.1 BAG4
## PYCR1.2 PYCR1
## LRRC58.2 LRRC58
## VIM-AS1 VIM-AS1
## THUMPD3-AS1.1 THUMPD3-AS1
## GLYR1.1 GLYR1
## CDK7 CDK7
## DDX51 DDX51
## AGPAT5 AGPAT5
## WASHC2C.1 WASHC2C
## NFKB2.1 NFKB2
## NSG1.1 NSG1
## MGST2.1 MGST2
## PRXL2B PRXL2B
## MFN2 MFN2
## CDS2.1 CDS2
## RSU1.1 RSU1
## FHIT.2 FHIT
## PLK4.2 PLK4
## LNPK LNPK
## KLF12 KLF12
## KCMF1.1 KCMF1
## CCDC69.1 CCDC69
## PPAT.1 PPAT
## HNRNPA1P48 HNRNPA1P48
## KLHDC7B KLHDC7B
## C2orf76 C2orf76
## TOX.2 TOX
## TTC19 TTC19
## PECR PECR
## ZNF227 ZNF227
## RELL2 RELL2
## MIS12.1 MIS12
## SLC35B3 SLC35B3
## TMEM267 TMEM267
## BCL10 BCL10
## INO80B INO80B
## NUP85.1 NUP85
## CCDC51 CCDC51
## ABCB7 ABCB7
## PDCD4-AS1 PDCD4-AS1
## S100B.2 S100B
## RFT1.1 RFT1
## PDE12.1 PDE12
## CD58.1 CD58
## ADD1.2 ADD1
## STK11.1 STK11
## CDCA3.2 CDCA3
## IWS1.1 IWS1
## CYTIP.2 CYTIP
## PCNX2 PCNX2
## B9D2 B9D2
## COX15 COX15
## PCBP1-AS1 PCBP1-AS1
## CAST.2 CAST
## HIF1A.1 HIF1A
## SLC43A1 SLC43A1
## CNEP1R1 CNEP1R1
## PRMT7.1 PRMT7
## ZNF263 ZNF263
## GNPNAT1.1 GNPNAT1
## NCAPH2.2 NCAPH2
## AC008892.1.1 AC008892.1
## LZTFL1.2 LZTFL1
## HUS1 HUS1
## TRNT1.1 TRNT1
## PIP4P2.1 PIP4P2
## PRKAG2 PRKAG2
## GM2A.1 GM2A
## NDFIP2.1 NDFIP2
## EHD1.1 EHD1
## PM20D2.1 PM20D2
## TMEM186 TMEM186
## CNOT10 CNOT10
## LRPPRC.2 LRPPRC
## AGL AGL
## TTC9C.1 TTC9C
## ALG13 ALG13
## BORA.1 BORA
## CIDEB.1 CIDEB
## WASHC2A WASHC2A
## NEU1.1 NEU1
## GNB1L GNB1L
## SMUG1.1 SMUG1
## VKORC1L1.1 VKORC1L1
## ATPAF2 ATPAF2
## KCTD17.1 KCTD17
## RCC1.2 RCC1
## DHRS4L2.1 DHRS4L2
## PRPF3.1 PRPF3
## TNFRSF4.2 TNFRSF4
## BLZF1.2 BLZF1
## CHIC2.1 CHIC2
## CORO7.2 CORO7
## FDX2.1 FDX2
## SATB1.1 SATB1
## USP11.2 USP11
## TFB1M TFB1M
## MRM1.1 MRM1
## NAA16 NAA16
## INAFM1.2 INAFM1
## TSPAN4.1 TSPAN4
## INSIG2 INSIG2
## BCS1L.1 BCS1L
## MED13L.1 MED13L
## GNGT2.2 GNGT2
## NSMF NSMF
## SRRD SRRD
## PTPN2.1 PTPN2
## HMGXB4.2 HMGXB4
## CALU.1 CALU
## SH2D3A.1 SH2D3A
## DELE1.1 DELE1
## TRIM65 TRIM65
## AC020915.2 AC020915.2
## PPP2R2D PPP2R2D
## LMO4.1 LMO4
## LIMK2.2 LIMK2
## DXO DXO
## CSNK2A1 CSNK2A1
## UNC93B1.1 UNC93B1
## CCNB2.2 CCNB2
## THOC1.2 THOC1
## BOD1 BOD1
## ITPK1.1 ITPK1
## GNA15.2 GNA15
## THAP12 THAP12
## NUP133.1 NUP133
## ZNF48 ZNF48
## ANAPC10 ANAPC10
## MED17 MED17
## AL136456.1.2 AL136456.1
## BAZ1A.1 BAZ1A
## PDE6B.1 PDE6B
## MOSMO.1 MOSMO
## FBXO22.1 FBXO22
## AL109840.2 AL109840.2
## CAMK2D.1 CAMK2D
## EGLN1 EGLN1
## SPTLC1 SPTLC1
## C3orf18 C3orf18
## UTP15.1 UTP15
## ZBTB48 ZBTB48
## MUTYH MUTYH
## JMJD1C.1 JMJD1C
## TMEM121 TMEM121
## STK32C STK32C
## METTL3 METTL3
## TTK.2 TTK
## GCSAM GCSAM
## EDEM1 EDEM1
## CUL5.1 CUL5
## ORC4 ORC4
## SLC25A12.1 SLC25A12
## CYHR1 CYHR1
## ECE1.2 ECE1
## TTC9 TTC9
## NUP155.2 NUP155
## RAF1.1 RAF1
## NVL NVL
## ID3.2 ID3
## FRAT2 FRAT2
## DOLPP1 DOLPP1
## CALCOCO1.2 CALCOCO1
## NKRF NKRF
## CCNH.1 CCNH
## MANBAL MANBAL
## WRAP53.1 WRAP53
## DENND6A DENND6A
## CSRNP1.1 CSRNP1
## GTSF1 GTSF1
## ACAD9 ACAD9
## PAFAH1B2 PAFAH1B2
## KAT5 KAT5
## ATP6V1C1 ATP6V1C1
## PTBP3.1 PTBP3
## KIF4A.2 KIF4A
## PHLDA2.1 PHLDA2
## PVT1 PVT1
## CDK11B.1 CDK11B
## STK19 STK19
## VOPP1.1 VOPP1
## NUP160.2 NUP160
## STT3B STT3B
## ASPHD2 ASPHD2
## ARHGAP27 ARHGAP27
## ZNF274 ZNF274
## GNPTAB.2 GNPTAB
## RBL1.2 RBL1
## PRPF18 PRPF18
## MON1A MON1A
## ABHD12.2 ABHD12
## FBXL12 FBXL12
## ZNF408 ZNF408
## ZNF714.2 ZNF714
## ARHGAP17.1 ARHGAP17
## ATP6V1A ATP6V1A
## CCAR2 CCAR2
## HIST2H2AC HIST2H2AC
## SLAMF6.2 SLAMF6
## SHLD1.1 SHLD1
## FBXO44.1 FBXO44
## NFX1 NFX1
## DVL2.1 DVL2
## C16orf58 C16orf58
## PTEN PTEN
## SNAPC1.1 SNAPC1
## CLU.1 CLU
## ZNF296.1 ZNF296
## RSPRY1 RSPRY1
## ERCC2.1 ERCC2
## MORC3 MORC3
## ELMOD2.1 ELMOD2
## BAP1 BAP1
## TTF2.2 TTF2
## ANKLE2 ANKLE2
## SWSAP1 SWSAP1
## SPINDOC.1 SPINDOC
## NCKAP1L.2 NCKAP1L
## CEP128.2 CEP128
## CDADC1 CDADC1
## DNAJC3.1 DNAJC3
## TOP1MT.1 TOP1MT
## TOM1 TOM1
## RSBN1L RSBN1L
## EIF2B4 EIF2B4
## UBL3.1 UBL3
## ZNF574 ZNF574
## ZBED1 ZBED1
## PTP4A3 PTP4A3
## CNP.2 CNP
## PALM2-AKAP2.2 PALM2-AKAP2
## BUB1B.2 BUB1B
## WDR62.2 WDR62
## SPSB2 SPSB2
## AFTPH AFTPH
## C1orf216.1 C1orf216
## RNF41 RNF41
## GSR.1 GSR
## HSPA14.1.1 HSPA14.1
## FCF1.1 FCF1
## BRD8 BRD8
## CCDC102A CCDC102A
## HARS2 HARS2
## STK17A.1 STK17A
## USP37.2 USP37
## SUV39H2.2 SUV39H2
## MFSD11 MFSD11
## FUBP3.1 FUBP3
## SHCBP1.2 SHCBP1
## SEC24C SEC24C
## CRYZL1 CRYZL1
## ATXN10 ATXN10
## TERF2 TERF2
## PHF10.1 PHF10
## TMEM43.1 TMEM43
## SPRYD7.1 SPRYD7
## CTU2 CTU2
## MAPK7 MAPK7
## APLP2.1 APLP2
## HVCN1.2 HVCN1
## KNSTRN.2 KNSTRN
## PXK.1 PXK
## FAAH2.2 FAAH2
## ARGLU1 ARGLU1
## RCC2.2 RCC2
## WDHD1.2 WDHD1
## PRORP PRORP
## TTF1.1 TTF1
## CERS4.1 CERS4
## CEP135.2 CEP135
## DONSON.2 DONSON
## LRRC45.2 LRRC45
## AKR1C3.1 AKR1C3
## HNRNPH1 HNRNPH1
## MAP7D3 MAP7D3
## KIF18B.2 KIF18B
## NT5E.1 NT5E
## TASOR.1 TASOR
## SRR.2 SRR
## SCO1 SCO1
## AC005842.1.2 AC005842.1
## TAF2.1 TAF2
## PCYOX1L.1 PCYOX1L
## MEMO1 MEMO1
## MED20 MED20
## ZNF862 ZNF862
## CCHCR1.1 CCHCR1
## HERPUD2.2 HERPUD2
## XKR8.1 XKR8
## PNMA1 PNMA1
## POC5.1 POC5
## FCHO1 FCHO1
## MICU2.1 MICU2
## DHX8 DHX8
## DNAAF5.1 DNAAF5
## MX1.2 MX1
## KCNN4 KCNN4
## BOLA1 BOLA1
## WDFY1.1 WDFY1
## PRR3 PRR3
## SGTB SGTB
## ARHGAP11A.2 ARHGAP11A
## CRCP CRCP
## CCNE1.2 CCNE1
## SERTAD3.1 SERTAD3
## QSOX1 QSOX1
## DOCK2.1 DOCK2
## MMP25-AS1.1 MMP25-AS1
## TNFRSF10A TNFRSF10A
## TRAPPC6B.1 TRAPPC6B
## STX2 STX2
## VRK2.1 VRK2
## ZNF195 ZNF195
## IL9R.1 IL9R
## TNPO1.1 TNPO1
## CLK2 CLK2
## C3orf14.1 C3orf14
## POLE2.2 POLE2
## CETN2 CETN2
## HSD17B7.1 HSD17B7
## HELLS.2 HELLS
## MHENCR.1 MHENCR
## DPH7 DPH7
## SLC25A17.1 SLC25A17
## AC008105.3.1 AC008105.3
## RNF6.1 RNF6
## TMEM68 TMEM68
## ASB6 ASB6
## ATP2B1-AS1 ATP2B1-AS1
## INPP4A.2 INPP4A
## TACC1.1 TACC1
## PTPN13.2 PTPN13
## EED.1 EED
## RBM48 RBM48
## ATG4C ATG4C
## SSX2IP SSX2IP
## MCOLN1 MCOLN1
## AKAP7 AKAP7
## ASL ASL
## NCBP1 NCBP1
## NUDT9 NUDT9
## ROGDI ROGDI
## POU2F2 POU2F2
## ZNF138.1 ZNF138
## ZBTB20.1 ZBTB20
## PDP1 PDP1
## FANCE.2 FANCE
## ELOA-AS1 ELOA-AS1
## CCM2.1 CCM2
## E2F2.2 E2F2
## GCC2.1 GCC2
## SOAT1.1 SOAT1
## MAFG.2 MAFG
## ITGA6.1 ITGA6
## E2F8.2 E2F8
## JUND.1 JUND
## TMPO-AS1.2 TMPO-AS1
## CES2 CES2
## TRAPPC13.1 TRAPPC13
## ZNF131 ZNF131
## TTLL12.2 TTLL12
## BCKDHB.1 BCKDHB
## CACNA2D4 CACNA2D4
## CYB561.1 CYB561
## ANKZF1 ANKZF1
## TIMM21.1 TIMM21
## E4F1.1 E4F1
## GPR183.2 GPR183
## CMPK2.2 CMPK2
## SLC15A4 SLC15A4
## ZNF626 ZNF626
## PCNT.1 PCNT
## GTPBP10 GTPBP10
## SIRT2 SIRT2
## TCHP TCHP
## PSMC3IP.2 PSMC3IP
## NUDT4.1 NUDT4
## CREG1.2 CREG1
## HIST1H3F.2 HIST1H3F
## NGLY1 NGLY1
## AANAT AANAT
## LARP4.1 LARP4
## GNS GNS
## PANK4 PANK4
## EIF5A2 EIF5A2
## NFRKB NFRKB
## FUZ FUZ
## ACTN1.2 ACTN1
## SCD.2 SCD
## LYRM7 LYRM7
## TRAV8-2.1 TRAV8-2
## TRG-AS1.2 TRG-AS1
## DDX28 DDX28
## PLPP1.2 PLPP1
## ZFP36L2.1 ZFP36L2
## CD3EAP.1 CD3EAP
## SLC38A1 SLC38A1
## XYLT1.1 XYLT1
## FASTKD5 FASTKD5
## UCK2.2 UCK2
## KCTD5 KCTD5
## MACO1 MACO1
## RAD18.2 RAD18
## FBXW2 FBXW2
## SMIM14 SMIM14
## ZNF12 ZNF12
## WDYHV1 WDYHV1
## SRP14-AS1 SRP14-AS1
## PIP4K2A.1 PIP4K2A
## RORA RORA
## ZNF641.1 ZNF641
## TMEM268 TMEM268
## MON1B MON1B
## RPUSD2 RPUSD2
## PRIM2.2 PRIM2
## EXOSC2.2 EXOSC2
## DERL3.1 DERL3
## AP5B1 AP5B1
## GFPT1.1 GFPT1
## MIEF1 MIEF1
## AGMAT AGMAT
## TAF1 TAF1
## MED18 MED18
## WDR43.1 WDR43
## DNAJA1 DNAJA1
## SCYL3 SCYL3
## ZNF224 ZNF224
## MAP2K7 MAP2K7
## RECQL4.2 RECQL4
## KIF3A KIF3A
## NECAP1 NECAP1
## VAT1 VAT1
## SCYL2 SCYL2
## TOP2B TOP2B
## POP1.1 POP1
## MBD6 MBD6
## B3GNT8.1 B3GNT8
## AP001011.1 AP001011.1
## TRAV16 TRAV16
## FBXL5 FBXL5
## CCDC66 CCDC66
## ABHD13 ABHD13
## DSTYK DSTYK
## TRIP6.2 TRIP6
## DNAJC1.2 DNAJC1
## TAF8 TAF8
## DSCC1.2 DSCC1
## PRKAR2A PRKAR2A
## PJA1 PJA1
## C12orf73 C12orf73
## AGK AGK
## TRGC2.2 TRGC2
## OFD1.1 OFD1
## EEF1AKMT1 EEF1AKMT1
## FIGNL1.2 FIGNL1
## AC010609.1.1 AC010609.1
## HDAC10 HDAC10
## PITPNM1 PITPNM1
## PIDD1.2 PIDD1
## SLC37A4.2 SLC37A4
## NECAB3.2 NECAB3
## RPUSD4 RPUSD4
## CRKL CRKL
## FKBP5.2 FKBP5
## AC083798.2 AC083798.2
## MR1 MR1
## FBXO4.2 FBXO4
## GGA2 GGA2
## SETD3 SETD3
## ZNF367.2 ZNF367
## PORCN.1 PORCN
## SLC35F6 SLC35F6
## YJU2 YJU2
## TMEM192 TMEM192
## TTLL1 TTLL1
## AP3M2.2 AP3M2
## VPS13B.1 VPS13B
## NUP214 NUP214
## CDC37L1 CDC37L1
## AC008875.3 AC008875.3
## SAYSD1 SAYSD1
## USF3 USF3
## CXorf38 CXorf38
## MTO1 MTO1
## METTL1.2 METTL1
## ENG ENG
## ACP2 ACP2
## MBD1 MBD1
## SP2 SP2
## CLCN3 CLCN3
## SLCO4A1.1 SLCO4A1
## SNX27 SNX27
## GPR18.2 GPR18
## RIF1.2 RIF1
## CD9.2 CD9
## PFAS.1 PFAS
## RETREG1.1 RETREG1
## ZNF84.1 ZNF84
## HSD17B12.2 HSD17B12
## NPTN.1 NPTN
## COG1 COG1
## CUL3 CUL3
## GPBP1 GPBP1
## MMS22L.2 MMS22L
## CYB5R4.1 CYB5R4
## ZC3H6 ZC3H6
## PPP2R5A PPP2R5A
## EZH1.1 EZH1
## WDR73 WDR73
## ERCC4 ERCC4
## ZNF668.2 ZNF668
## IL23A.1 IL23A
## TMEM87A TMEM87A
## USF1.1 USF1
## GTF2B GTF2B
## ZNF738.1 ZNF738
## SPC24.2 SPC24
## DLGAP4 DLGAP4
## PI4KB PI4KB
## VTI1A.1 VTI1A
## NAT14 NAT14
## KLHL7 KLHL7
## TRBV7-9.2 TRBV7-9
## FASTKD1 FASTKD1
## UBA7 UBA7
## CCND3.1 CCND3
## DNAJC10 DNAJC10
## AIG1.2 AIG1
## LINC02611 LINC02611
## PDIA5.1 PDIA5
## FBXL14 FBXL14
## NAGA.2 NAGA
## NR4A2.2 NR4A2
## CANT1 CANT1
## PPCDC.1 PPCDC
## RPP25.2 RPP25
## GADD45B.2 GADD45B
## C6orf62 C6orf62
## VCPKMT VCPKMT
## ACBD6 ACBD6
## AFG3L2.2 AFG3L2
## FCGRT.1 FCGRT
## LTBP4 LTBP4
## AGFG2 AGFG2
## STX7 STX7
## PAPOLG PAPOLG
## NDEL1 NDEL1
## HECTD3 HECTD3
## PKNOX1 PKNOX1
## RNF13 RNF13
## MTG2.1 MTG2
## C11orf21.2 C11orf21
## CSPP1.2 CSPP1
## CLP1 CLP1
## GPBP1L1 GPBP1L1
## C21orf58.2 C21orf58
## UBA6 UBA6
## CDC25A.2 CDC25A
## SUPT6H SUPT6H
## NBPF14.1 NBPF14
## GHDC.1 GHDC
## SEC14L1 SEC14L1
## KLHL12 KLHL12
## DNAJB9 DNAJB9
## CCP110.1 CCP110
## EIF4E3.1 EIF4E3
## MAP3K11 MAP3K11
## ERO1A.1 ERO1A
## ODF3B.2 ODF3B
## FEM1B FEM1B
## TRIP11 TRIP11
## ZWILCH.2 ZWILCH
## CRAT CRAT
## NEMF.1 NEMF
## EFL1.1 EFL1
## MCAT.1 MCAT
## TRBV28 TRBV28
## SPINK2.2 SPINK2
## NFYA NFYA
## DLEU1 DLEU1
## DNAJC11 DNAJC11
## TAF11.1 TAF11
## MCEE MCEE
## BBS7 BBS7
## SHQ1 SHQ1
## ARHGAP5 ARHGAP5
## TOR1AIP2 TOR1AIP2
## IER5L.2 IER5L
## ITFG2 ITFG2
## AC111182.1.1 AC111182.1
## WDR83.1 WDR83
## SPTY2D1 SPTY2D1
## RCBTB2.1 RCBTB2
## SMAP1.1 SMAP1
## COQ10B COQ10B
## BISPR.1 BISPR
## SNRNP48 SNRNP48
## MRPL1.1 MRPL1
## AL136454.1 AL136454.1
## CUL4A CUL4A
## HIBADH.1 HIBADH
## DTD2 DTD2
## GRPEL2 GRPEL2
## GABPA GABPA
## ING2.1 ING2
## OGDH OGDH
## TROAP.2 TROAP
## MRI1.1 MRI1
## AC139720.1.2 AC139720.1
## MTFR2.2 MTFR2
## FLNB.1 FLNB
## POLH.1 POLH
## VARS.1 VARS
## TUBGCP3.1 TUBGCP3
## CDPF1.1 CDPF1
## VSIG1.1 VSIG1
## MTX2.1 MTX2
## DPYD-AS1.2 DPYD-AS1
## BCL2L1.2 BCL2L1
## SCAF11 SCAF11
## POR POR
## DBN1.2 DBN1
## RNF125.1 RNF125
## ZNF550 ZNF550
## PCSK7.1 PCSK7
## NEMP2.1 NEMP2
## ATXN3 ATXN3
## LINC01184.2 LINC01184
## HERC1 HERC1
## ENTPD6 ENTPD6
## PIK3R2 PIK3R2
## GZMH.2 GZMH
## AC119396.1.1 AC119396.1
## RHOT1.1 RHOT1
## XRCC2.2 XRCC2
## CCND2.1 CCND2
## PACC1 PACC1
## SLC25A51 SLC25A51
## IRF2BP2 IRF2BP2
## C6orf120 C6orf120
## GGCX.1 GGCX
## MTRR MTRR
## BCL2L13 BCL2L13
## ZNF160 ZNF160
## PSMA3-AS1 PSMA3-AS1
## STX6 STX6
## FAM118A.1 FAM118A
## UXS1.1 UXS1
## PUS1.1 PUS1
## HES6.2 HES6
## VPS45 VPS45
## CPSF6 CPSF6
## FIZ1.1 FIZ1
## LINC00426 LINC00426
## EID3 EID3
## EMILIN2.1 EMILIN2
## CCNT1.1 CCNT1
## ZNF346 ZNF346
## EFCAB11.2 EFCAB11
## ENOSF1.2 ENOSF1
## RIOX1 RIOX1
## CUL2 CUL2
## PSD4.2 PSD4
## VILL VILL
## MED21.1 MED21
## DIS3 DIS3
## KRBOX4 KRBOX4
## TGOLN2.1 TGOLN2
## HGS HGS
## LRRC40.2 LRRC40
## PYGO2 PYGO2
## CABIN1 CABIN1
## RNF44 RNF44
## SH3BP5L SH3BP5L
## BTN2A1 BTN2A1
## FAM189B FAM189B
## C4orf46.2 C4orf46
## ZNF302.1 ZNF302
## TOMM34.1 TOMM34
## RNFT1 RNFT1
## SNHG12 SNHG12
## TBK1.1 TBK1
## EEPD1 EEPD1
## WDR86-AS1.1 WDR86-AS1
## LRRCC1.2 LRRCC1
## MCCC1 MCCC1
## HYI.2 HYI
## C7orf25 C7orf25
## TPM4.2 TPM4
## ZNF692 ZNF692
## UBE2K UBE2K
## BAZ2A BAZ2A
## C3orf38 C3orf38
## MTA1.1 MTA1
## ZNF683.2 ZNF683
## UPRT UPRT
## ODC1 ODC1
## USP21 USP21
## PBK.2 PBK
## ZFP90 ZFP90
## RILP.1 RILP
## STRADA STRADA
## MSANTD3 MSANTD3
## PAX8-AS1 PAX8-AS1
## PCF11 PCF11
## LONP1.2 LONP1
## SGSH.2 SGSH
## LINS1.1 LINS1
## OSBPL3.1 OSBPL3
## SLC30A9 SLC30A9
## RAD54B.2 RAD54B
## FAM24B FAM24B
## ADAT3.1 ADAT3
## TTI1.1 TTI1
## APTR APTR
## TMEM185A TMEM185A
## NFATC2IP.1 NFATC2IP
## THAP3 THAP3
## RNMT RNMT
## VAPB.1 VAPB
## C5orf56 C5orf56
## ZNF264 ZNF264
## MXRA7.1 MXRA7
## TYK2 TYK2
## YAE1 YAE1
## CEP97.1 CEP97
## NSDHL.1 NSDHL
## NHEJ1 NHEJ1
## SNRNP200 SNRNP200
## UGDH UGDH
## ZMYM6 ZMYM6
## SLC38A2 SLC38A2
## UBQLN1 UBQLN1
## BIK BIK
## FOSB FOSB
## PATZ1 PATZ1
## PRR34-AS1 PRR34-AS1
## MED7 MED7
## TCF3.1 TCF3
## SMARCA4.1 SMARCA4
## EAF1.2 EAF1
## NNT-AS1 NNT-AS1
## APOOL APOOL
## ZNF611 ZNF611
## GRK2.1 GRK2
## MAK16 MAK16
## PPP1R21 PPP1R21
## ARAP2.2 ARAP2
## RTL6 RTL6
## FKBP2 FKBP2
## PHIP.1 PHIP
## IL3RA IL3RA
## AGAP2 AGAP2
## INTS9 INTS9
## ADAM8.2 ADAM8
## ADAR ADAR
## WDR89 WDR89
## C9orf85 C9orf85
## DEPDC1B.2 DEPDC1B
## ZNF426 ZNF426
## PCED1B.2 PCED1B
## TXNRD2.1 TXNRD2
## TMCO6 TMCO6
## DIABLO DIABLO
## FHL3 FHL3
## RNF149.1 RNF149
## NFATC1.1 NFATC1
## TTC5 TTC5
## MRE11.1 MRE11
## LARP1.2 LARP1
## SMAD4 SMAD4
## NSUN2 NSUN2
## ZUP1 ZUP1
## LUC7L2.1 LUC7L2
## SNIP1.1 SNIP1
## TEDC2.2 TEDC2
## B4GAT1 B4GAT1
## CTC1 CTC1
## NUFIP1 NUFIP1
## SPATS2.1 SPATS2
## PPM1A PPM1A
## SAMD3.2 SAMD3
## ZNF226 ZNF226
## CCDC9 CCDC9
## SLC25A45 SLC25A45
## LTB4R2.1 LTB4R2
## XPO5 XPO5
## CSF1.2 CSF1
## RAB34 RAB34
## RCAN1 RCAN1
## SLC18B1 SLC18B1
## IZUMO4 IZUMO4
## MOSPD2 MOSPD2
## RIBC2.2 RIBC2
## SMC5.1 SMC5
## ZNF7.1 ZNF7
## RPE RPE
## SCPEP1.1 SCPEP1
## SLC27A3.1 SLC27A3
## HEATR6 HEATR6
## RNF135 RNF135
## PRIMPOL.1 PRIMPOL
## NAT9 NAT9
## TRAV17.1 TRAV17
## AP000787.1.1 AP000787.1
## RYK RYK
## ARHGEF1 ARHGEF1
## PLK1.2 PLK1
## ERGIC1.1 ERGIC1
## PPM1D PPM1D
## AMPD2 AMPD2
## NAPA-AS1 NAPA-AS1
## ZNF397 ZNF397
## PIK3R1.1 PIK3R1
## FKBP15.1 FKBP15
## ZEB1.1 ZEB1
## TM9SF4 TM9SF4
## TNFRSF18.1 TNFRSF18
## NFKBIA.1 NFKBIA
## SHOC2 SHOC2
## IPO4.1 IPO4
## VSIG8 VSIG8
## ARHGEF2 ARHGEF2
## TNFAIP3 TNFAIP3
## VEZT VEZT
## AASDH.1 AASDH
## WDR6 WDR6
## B4GALT7 B4GALT7
## PARP11.2 PARP11
## GTF3C3 GTF3C3
## COQ10A COQ10A
## CTPS1.2 CTPS1
## CA5B CA5B
## SAP130.1 SAP130
## FAM120A FAM120A
## RNF2 RNF2
## UBAP2.1 UBAP2
## PCBP4.1 PCBP4
## UFSP2 UFSP2
## TC2N.2 TC2N
## CCDC14.1 CCDC14
## UBE4A UBE4A
## ZBTB4 ZBTB4
## ZNF33B.1 ZNF33B
## HELZ HELZ
## WDR4.2 WDR4
## SEC23A.1 SEC23A
## APTX APTX
## MALT1 MALT1
## FECH.1 FECH
## GABARAPL1.2 GABARAPL1
## INTS3 INTS3
## LY9 LY9
## TRAV8-4 TRAV8-4
## TADA2A.2 TADA2A
## ELAC1 ELAC1
## ZNF322 ZNF322
## EEF1AKMT4 EEF1AKMT4
## MARK2 MARK2
## ZDHHC7 ZDHHC7
## ATP2A1-AS1.2 ATP2A1-AS1
## PDCD11 PDCD11
## IDE IDE
## TADA2B TADA2B
## KIF14.2 KIF14
## ACYP2 ACYP2
## IPO8 IPO8
## RBM10 RBM10
## CDC16 CDC16
## FANCM.2 FANCM
## XPO7.1 XPO7
## HCG18 HCG18
## UEVLD.1 UEVLD
## FAM126A FAM126A
## AC125494.1 AC125494.1
## POLR3H.2 POLR3H
## ZBTB14 ZBTB14
## ZBED5-AS1 ZBED5-AS1
## UBASH3A.1 UBASH3A
## SPPL2A.1 SPPL2A
## EIPR1.1 EIPR1
## TIMM44.1 TIMM44
## SLC35A1 SLC35A1
## TRMO TRMO
## TXNL4B TXNL4B
## LHFPL5 LHFPL5
## SNX12 SNX12
## GMFB GMFB
## PCYOX1 PCYOX1
## ZC3HC1.1 ZC3HC1
## PPOX PPOX
## SNTA1.1 SNTA1
## AC017083.2 AC017083.2
## MYPOP MYPOP
## CCDC130 CCDC130
## COQ8B COQ8B
## MEPCE MEPCE
## DPP9 DPP9
## SCFD1 SCFD1
## SERPINB9.1 SERPINB9
## DCTN4 DCTN4
## CEP295.2 CEP295
## C11orf71 C11orf71
## HMMR.2 HMMR
## INTS7.1 INTS7
## FRMD8 FRMD8
## MOB3C MOB3C
## BCDIN3D BCDIN3D
## GPAT4 GPAT4
## STRADB STRADB
## DDX55.2 DDX55
## KIF9.1 KIF9
## MSRA.1 MSRA
## ZNF85.1 ZNF85
## PHYH PHYH
## NARF NARF
## TP53BP1 TP53BP1
## SCRN3 SCRN3
## TCAIM TCAIM
## TCEAL4.1 TCEAL4
## TCOF1.1 TCOF1
## ZSCAN32 ZSCAN32
## NMB.1 NMB
## SLC25A19.2 SLC25A19
## TICAM1 TICAM1
## CEP290 CEP290
## IQCB1.1 IQCB1
## PRKDC.2 PRKDC
## CAMK2N1.1 CAMK2N1
## APOBEC3F APOBEC3F
## KATNAL1 KATNAL1
## ITPR2 ITPR2
## WDR74 WDR74
## ANKRD9.2 ANKRD9
## MAN2B2 MAN2B2
## IRF2BP1 IRF2BP1
## FASTKD3 FASTKD3
## C2CD2L.1 C2CD2L
## HOMEZ HOMEZ
## TBC1D15 TBC1D15
## PRKCD PRKCD
## METTL6 METTL6
## FAAP24.2 FAAP24
## HRH2.2 HRH2
## C1GALT1.1 C1GALT1
## R3HDM1.1 R3HDM1
## ZNF280D ZNF280D
## AKAP17A AKAP17A
## RANBP6 RANBP6
## ODR4.1 ODR4
## AC004797.1 AC004797.1
## RNF139 RNF139
## DDX11.2 DDX11
## POT1 POT1
## RAD51D.1 RAD51D
## NUP43.1 NUP43
## CRTC3.1 CRTC3
## POM121C POM121C
## LINC00476 LINC00476
## NFE2L3.2 NFE2L3
## SP3.1 SP3
## SLC25A28 SLC25A28
## ARHGEF6.1 ARHGEF6
## C16orf74.2 C16orf74
## CHD7.1 CHD7
## IL6ST.2 IL6ST
## PRKCI PRKCI
## GOLPH3L GOLPH3L
## PPP3R1.1 PPP3R1
## NUP210.2 NUP210
## GEMIN2.1 GEMIN2
## C17orf58.1 C17orf58
## GRIPAP1.1 GRIPAP1
## LINC01572.2 LINC01572
## ZNF71 ZNF71
## ATG16L2 ATG16L2
## TGFBRAP1 TGFBRAP1
## DHX16.1 DHX16
## HIGD1A HIGD1A
## GUF1 GUF1
## PIGU.2 PIGU
## NUP205.2 NUP205
## PCYT1A.1 PCYT1A
## CNOT2 CNOT2
## FBXL17.1 FBXL17
## NEK9 NEK9
## PLAGL2.1 PLAGL2
## TMEM63B TMEM63B
## MLLT11.1 MLLT11
## NUDT15.1 NUDT15
## ACADSB ACADSB
## AP002360.1 AP002360.1
## LPCAT3 LPCAT3
## SNRPD3.1 SNRPD3
## NDUFAF6.1 NDUFAF6
## ATRIP.1 ATRIP
## GFOD2 GFOD2
## CSNK1D CSNK1D
## RANBP3.1 RANBP3
## CCDC138.1 CCDC138
## TRAV29DV5.1 TRAV29DV5
## ALG6 ALG6
## PUM3.1 PUM3
## NACC1.1 NACC1
## ALKBH4 ALKBH4
## MICALL1 MICALL1
## PBX2 PBX2
## HYAL3 HYAL3
## NT5C2 NT5C2
## AL391056.1.1 AL391056.1
## ACIN1 ACIN1
## BEST1 BEST1
## TARS2.1 TARS2
## RNF26 RNF26
## SIN3B SIN3B
## POLR2J3 POLR2J3
## LMBR1L LMBR1L
## AC005332.4 AC005332.4
## HIST1H3G.2 HIST1H3G
## EIF4G3.1 EIF4G3
## FBXO8 FBXO8
## TUBGCP4 TUBGCP4
## SENP3 SENP3
## ZC3H11A ZC3H11A
## C9orf135.2 C9orf135
## REG4.1 REG4
## GABPB1-AS1 GABPB1-AS1
## TONSL.2 TONSL
## TRMT13 TRMT13
## CENPQ.2 CENPQ
## ABITRAM ABITRAM
## MASTL.2 MASTL
## SULT1A1.1 SULT1A1
## AC016747.1.2 AC016747.1
## GKAP1.2 GKAP1
## USPL1 USPL1
## AC010894.3.1 AC010894.3
## LIG4 LIG4
## KPNA6.1 KPNA6
## ZGRF1.2 ZGRF1
## POLA1.2 POLA1
## ZNF569 ZNF569
## WDR33.1 WDR33
## ZNF586 ZNF586
## PSTK PSTK
## ZNF836 ZNF836
## AC016876.1 AC016876.1
## FMR1 FMR1
## JADE2 JADE2
## CUL4B CUL4B
## ZC4H2 ZC4H2
## GPATCH3 GPATCH3
## ARHGAP45.1 ARHGAP45
## LXN.1 LXN
## PPIL2.1 PPIL2
## USP33 USP33
## TBC1D1.1 TBC1D1
## FXYD2.1 FXYD2
## DHX58 DHX58
## ZG16B ZG16B
## ZBTB2.1 ZBTB2
## NCALD.1 NCALD
## IFNGR1.2 IFNGR1
## KCTD9.1 KCTD9
## AL135818.1.2 AL135818.1
## SPTBN1.2 SPTBN1
## PDIK1L.1 PDIK1L
## IRAK1BP1 IRAK1BP1
## SLC27A2.1 SLC27A2
## C1orf112.2 C1orf112
## RAD17 RAD17
## NAA15.1 NAA15
## ZNF789.1 ZNF789
## TPP2.1 TPP2
## CEP83.1 CEP83
## AAK1 AAK1
## GEMIN5 GEMIN5
## TRIM13 TRIM13
## QTRT2 QTRT2
## ZNF257.2 ZNF257
## PPP2R1B.2 PPP2R1B
## CLASRP CLASRP
## TBC1D22A TBC1D22A
## SREK1 SREK1
## LLGL1 LLGL1
## PIP4K2C PIP4K2C
## EXOC1 EXOC1
## FXYD7.1 FXYD7
## IDNK IDNK
## TTC31 TTC31
## INSIG1 INSIG1
## TRBV7-2 TRBV7-2
## ZNF592 ZNF592
## HINT3.1 HINT3
## PAXIP1-AS2 PAXIP1-AS2
## PNPLA6 PNPLA6
## CPT1A.2 CPT1A
## CASTOR2 CASTOR2
## NFE2L2 NFE2L2
## RMND1 RMND1
## STAG2.1 STAG2
## PTCD1 PTCD1
## LIN7C LIN7C
## ZDHHC5.1 ZDHHC5
## SACM1L.1 SACM1L
## ABCB10.1 ABCB10
## ENDOV ENDOV
## VPS11 VPS11
## CDC25C.2 CDC25C
## RBFOX2 RBFOX2
## POMT1.1 POMT1
## SLC35A2 SLC35A2
## EMC1 EMC1
## RNF121.2 RNF121
## MFSD14A MFSD14A
## ZFYVE19.1 ZFYVE19
## AFMID.2 AFMID
## CHUK CHUK
## SNHG10 SNHG10
## SAMD9L.1 SAMD9L
## ATP8B2.1 ATP8B2
## OXR1 OXR1
## ZNF174 ZNF174
## ZMYM1 ZMYM1
## MBNL3.1 MBNL3
## B4GALT2 B4GALT2
## ANKRD44-IT1 ANKRD44-IT1
## TRAV8-6 TRAV8-6
## YTHDC1.1 YTHDC1
## GPR68.1 GPR68
## CRELD1 CRELD1
## STAT5B STAT5B
## COLGALT1.1 COLGALT1
## ARRDC1-AS1 ARRDC1-AS1
## CEP152.2 CEP152
## STAT3 STAT3
## ABHD11.1 ABHD11
## TOMM40L.2 TOMM40L
## CEP72.2 CEP72
## RPRD2 RPRD2
## FAM3C.1 FAM3C
## WASHC4 WASHC4
## LINC02084.1 LINC02084
## PGPEP1 PGPEP1
## PUS3 PUS3
## CEP85.2 CEP85
## AC245014.3.1 AC245014.3
## TRBV9.2 TRBV9
## NFIC.1 NFIC
## ZBTB38.1 ZBTB38
## SPRTN SPRTN
## GPN2.1 GPN2
## RANBP9.1 RANBP9
## TGFBR3L TGFBR3L
## ANAPC4 ANAPC4
## ADGRE1.2 ADGRE1
## TRAPPC2 TRAPPC2
## CHPF2 CHPF2
## ESPL1.2 ESPL1
## FANCL.2 FANCL
## ZSCAN21 ZSCAN21
## EZH2.2 EZH2
## NSD2.2 NSD2
## AKAP1.1 AKAP1
## ARMC9 ARMC9
## EXOG EXOG
## SENP6 SENP6
## PRDM4.1 PRDM4
## RNPC3 RNPC3
## SMPD2 SMPD2
## ENDOD1.1 ENDOD1
## RETREG3 RETREG3
## HSF2.1 HSF2
## TRMU.1 TRMU
## GLA.1 GLA
## SPTLC2.2 SPTLC2
## CBR4 CBR4
## CBY1 CBY1
## IQCG IQCG
## VNN2.1 VNN2
## PRKCQ.1 PRKCQ
## TRAV35 TRAV35
## PAK1 PAK1
## RSF1.1 RSF1
## FBXO28 FBXO28
## TRAV4 TRAV4
## ZNF672 ZNF672
## PPP3CC.2 PPP3CC
## SESN3.2 SESN3
## LIN52.2 LIN52
## CCDC174 CCDC174
## FAM53C FAM53C
## NEIL2.1 NEIL2
## NUDT7 NUDT7
## CREB1 CREB1
## ZNF250 ZNF250
## NCDN.1 NCDN
## SMG8 SMG8
## ZNF41.1 ZNF41
## BTD BTD
## GADD45G.2 GADD45G
## PARP6 PARP6
## ASXL1.1 ASXL1
## CAPN7.1 CAPN7
## ZNF281 ZNF281
## NARS2.1 NARS2
## SERPINH1.1 SERPINH1
## TRBV2.2 TRBV2
## AC127521.1.1 AC127521.1
## STXBP1 STXBP1
## PRKRIP1 PRKRIP1
## MXI1 MXI1
## STOML1.1 STOML1
## TRAV27.1 TRAV27
## GOLGA8A.1 GOLGA8A
## TTC14 TTC14
## DDX59 DDX59
## IL15RA.2 IL15RA
## FEM1A FEM1A
## TOR1B TOR1B
## KIAA1958 KIAA1958
## SLC25A40.1 SLC25A40
## ZNF75A ZNF75A
## ITK.1 ITK
## ASB16-AS1 ASB16-AS1
## C5orf63 C5orf63
## C11orf98 C11orf98
## BOD1L1.1 BOD1L1
## DDX60 DDX60
## CEP120 CEP120
## SMKR1 SMKR1
## CENPA.2 CENPA
## FBLN7.2 FBLN7
## TMEM44-AS1 TMEM44-AS1
## TRIM14.1 TRIM14
## CHEK2.2 CHEK2
## PCNX1.1 PCNX1
## TMEM185B TMEM185B
## PHC1 PHC1
## ZNF567 ZNF567
## MED9 MED9
## NDE1 NDE1
## CEP57L1.1 CEP57L1
## PCMTD1 PCMTD1
## COG8 COG8
## AVEN AVEN
## LRRFIP2.1 LRRFIP2
## GNG2.1 GNG2
## EXT2 EXT2
## XPNPEP3 XPNPEP3
## BIRC6 BIRC6
## MTHFD1L.2 MTHFD1L
## FZR1.1 FZR1
## ZNF3 ZNF3
## CD302.1 CD302
## ZNF487 ZNF487
## MANEA-DT MANEA-DT
## SCARB2.1 SCARB2
## HASPIN.2 HASPIN
## MTMR6 MTMR6
## PEX6 PEX6
## FAM111A-DT FAM111A-DT
## AKAP8 AKAP8
## SAMD4B SAMD4B
## CBX6.1 CBX6
## SRF SRF
## TRAV26-1 TRAV26-1
## TENT4B TENT4B
## KCTD13 KCTD13
## HS1BP3 HS1BP3
## HACL1 HACL1
## NLRX1.1 NLRX1
## MECOM MECOM
## AL662797.1.2 AL662797.1
## RBM28.1 RBM28
## MINDY2 MINDY2
## AURKA.2 AURKA
## GEN1.2 GEN1
## CASC4 CASC4
## FTX FTX
## CXorf40A CXorf40A
## MFSD3 MFSD3
## TRAPPC12 TRAPPC12
## CLCC1.1 CLCC1
## TBC1D9B.1 TBC1D9B
## MARF1 MARF1
## PEX10 PEX10
## ZFP36.2 ZFP36
## NDST2 NDST2
## PCMTD2 PCMTD2
## CDK11A CDK11A
## PARP2.1 PARP2
## PKIA.1 PKIA
## MEX3C MEX3C
## BCL9L.2 BCL9L
## TRIM24.2 TRIM24
## AP003086.1.1 AP003086.1
## DIPK2A DIPK2A
## OSM.2 OSM
## PLEKHA1.2 PLEKHA1
## PDSS1.2 PDSS1
## SH3BP2.1 SH3BP2
## ATF5.1 ATF5
## VIPAS39 VIPAS39
## KRT1.2 KRT1
## MTMR2.1 MTMR2
## SETDB1 SETDB1
## WDR11 WDR11
## ZC3H3.1 ZC3H3
## LINC02245.1 LINC02245
## IFT74.1 IFT74
## HMGXB3 HMGXB3
## CHI3L2.2 CHI3L2
## CCDC93.1 CCDC93
## P2RX5.2 P2RX5
## SUPT5H SUPT5H
## DYNLRB1 DYNLRB1
## SREBF1.2 SREBF1
## TLR9 TLR9
## KIF1C.1 KIF1C
## UBE4B.1 UBE4B
## SLC17A5 SLC17A5
## TRAPPC11 TRAPPC11
## DHX33.2 DHX33
## RENBP RENBP
## UBXN2B UBXN2B
## TMEM41B TMEM41B
## TRMT1L TRMT1L
## TRAV23DV6 TRAV23DV6
## UGCG UGCG
## CAPN12.2 CAPN12
## ITGA5 ITGA5
## FOXP3 FOXP3
## EFR3A EFR3A
## MPZL3.1 MPZL3
## TMEM71.1 TMEM71
## ZNF747 ZNF747
## SFSWAP SFSWAP
## PRKCA-AS1.1 PRKCA-AS1
## ZNF213-AS1 ZNF213-AS1
## ADAT2 ADAT2
## WWOX.2 WWOX
## NORAD NORAD
## EDC4.1 EDC4
## SCAMP1-AS1 SCAMP1-AS1
## ZNF362 ZNF362
## ZNF579 ZNF579
## AC009779.2 AC009779.2
## TCEANC2.2 TCEANC2
## ZNF568 ZNF568
## FAM174A FAM174A
## IGHMBP2 IGHMBP2
## GIGYF2.1 GIGYF2
## PPRC1.1 PPRC1
## MED27.1 MED27
## ACSS1.1 ACSS1
## TMEM63A.2 TMEM63A
## SIRT3 SIRT3
## FAM227B FAM227B
## AXIN1.1 AXIN1
## ADAMTS6 ADAMTS6
## CHD1 CHD1
## IER3.1 IER3
## GTF2E1 GTF2E1
## AC005562.1 AC005562.1
## ZNF197 ZNF197
## AC092652.2 AC092652.2
## MIGA2 MIGA2
## SLAIN1 SLAIN1
## CDK16 CDK16
## CREB3L2.1 CREB3L2
## GRAP.1 GRAP
## CTNS CTNS
## AP001347.1.2 AP001347.1
## ITPRID2.1 ITPRID2
## THAP7-AS1.1 THAP7-AS1
## INPP5D.1 INPP5D
## TDP1.1 TDP1
## LY6G5C LY6G5C
## MSANTD4 MSANTD4
## HIBCH HIBCH
## SLC9B2 SLC9B2
## AL050341.2 AL050341.2
## CDKN1A.1 CDKN1A
## MORN2.1 MORN2
## C19orf54 C19orf54
## D2HGDH D2HGDH
## TMEM38B.2 TMEM38B
## COP1.1 COP1
## ADPRM ADPRM
## MTBP.2 MTBP
## SLC46A3 SLC46A3
## CTDSPL2.1 CTDSPL2
## TFDP2.1 TFDP2
## ST3GAL3.1 ST3GAL3
## AC023424.3 AC023424.3
## ZNF600 ZNF600
## KPNA5 KPNA5
## ALG10 ALG10
## PIGQ PIGQ
## SGK3 SGK3
## TNFAIP1 TNFAIP1
## LTN1 LTN1
## RAP2C RAP2C
## ZNF350 ZNF350
## VIRMA VIRMA
## HERC6 HERC6
## DHX38 DHX38
## CCDC125 CCDC125
## AC244090.1.1 AC244090.1
## NXPE3 NXPE3
## DNAJC14 DNAJC14
## UNC13D.1 UNC13D
## SPACA9 SPACA9
## ICE2.2 ICE2
## TMEM177 TMEM177
## LRFN4 LRFN4
## ZNF518A ZNF518A
## ZBTB25 ZBTB25
## TSPOAP1-AS1 TSPOAP1-AS1
## FGFBP2.2 FGFBP2
## SHISAL2A SHISAL2A
## NDUFAF7 NDUFAF7
## VAV1 VAV1
## FAM234A.1 FAM234A
## KIZ KIZ
## SLC5A6 SLC5A6
## ZSWIM9 ZSWIM9
## ELP1 ELP1
## LRRC57.1 LRRC57
## ARID1A.1 ARID1A
## CPOX.1 CPOX
## TAF3 TAF3
## YME1L1 YME1L1
## SIRT6 SIRT6
## PLEKHG3.1 PLEKHG3
## YWHAG.2 YWHAG
## BANP BANP
## PIK3R4 PIK3R4
## ZMYM4.1 ZMYM4
## JPX JPX
## TICRR.2 TICRR
## LINC01215 LINC01215
## AP4M1.1 AP4M1
## GALNT7.1 GALNT7
## RNASEL RNASEL
## ZNF317 ZNF317
## FICD.1 FICD
## NEMP1.2 NEMP1
## METTL2A METTL2A
## ATPSCKMT.1 ATPSCKMT
## RUFY3.1 RUFY3
## MZF1-AS1 MZF1-AS1
## TRAV13-2 TRAV13-2
## ZNF764.1 ZNF764
## NHLRC3 NHLRC3
## XPO1.1 XPO1
## SPAST SPAST
## PEX3.1 PEX3
## DYRK1A.1 DYRK1A
## SUGP1 SUGP1
## SEPTIN11.1 SEPTIN11
## TMEM184C TMEM184C
## SPHK2 SPHK2
## MAP3K12 MAP3K12
## WDR75.1 WDR75
## DHRS7B.2 DHRS7B
## PIGO PIGO
## ANKRD26.1 ANKRD26
## RBM15.1 RBM15
## TRGV3.2 TRGV3
## MAP9 MAP9
## TFEB TFEB
## PLCD1 PLCD1
## AP4B1 AP4B1
## TRAV9-2 TRAV9-2
## TRAV12-3 TRAV12-3
## TIAM1.2 TIAM1
## LEF1-AS1.1 LEF1-AS1
## DTX2 DTX2
## ZNF124 ZNF124
## NIFK-AS1 NIFK-AS1
## ESF1.1 ESF1
## SPPL2B.1 SPPL2B
## PIAS3 PIAS3
## LINC01137 LINC01137
## TRMT5 TRMT5
## RFC3.2 RFC3
## PHF2 PHF2
## ZNF100.1 ZNF100
## GPRIN3.1 GPRIN3
## PLEKHA7 PLEKHA7
## PURB PURB
## SSBP3.1 SSBP3
## KNOP1 KNOP1
## ARID4A ARID4A
## HEATR3 HEATR3
## CNTROB.1 CNTROB
## IPO9.2 IPO9
## GINS1.2 GINS1
## PLD6 PLD6
## RABL2B RABL2B
## SMIM13.1 SMIM13
## ZCCHC3 ZCCHC3
## FGFR1.1 FGFR1
## CEP44 CEP44
## ZNF445 ZNF445
## HPS5 HPS5
## ABHD4 ABHD4
## POM121.1 POM121
## LMAN2L LMAN2L
## RPP40 RPP40
## BSDC1 BSDC1
## GNL3L.1 GNL3L
## THAP5 THAP5
## DTX3 DTX3
## ATG5 ATG5
## PPP4R1 PPP4R1
## ZKSCAN1 ZKSCAN1
## AC015871.1.1 AC015871.1
## TRAV12-1 TRAV12-1
## ZNF10 ZNF10
## MX2.2 MX2
## SENP5.1 SENP5
## ACOX1 ACOX1
## ZNF276.1 ZNF276
## NAXD NAXD
## ZNF33A ZNF33A
## NET1.2 NET1
## KCTD6 KCTD6
## LINC02361 LINC02361
## C1orf109 C1orf109
## ASCC3 ASCC3
## RIPK3.1 RIPK3
## SON SON
## ZFAND5 ZFAND5
## NAPEPLD.1 NAPEPLD
## WIPI1.1 WIPI1
## SMAGP SMAGP
## BPGM.1 BPGM
## TOP3A.2 TOP3A
## C10orf88 C10orf88
## KANSL2.1 KANSL2
## KLF16 KLF16
## COMMD10.1 COMMD10
## STXBP3 STXBP3
## MAP1LC3B2 MAP1LC3B2
## MMS19.1 MMS19
## GTF3C4 GTF3C4
## AL157938.2 AL157938.2
## NINJ2.1 NINJ2
## GSEC GSEC
## SLC12A9 SLC12A9
## NDRG3.1 NDRG3
## ZNF407.1 ZNF407
## PIGW.1 PIGW
## LYZ.2 LYZ
## CEP85L.1 CEP85L
## SUCO.2 SUCO
## PRMT5-AS1 PRMT5-AS1
## HIST1H1E HIST1H1E
## PTPRN2.1 PTPRN2
## MECP2.1 MECP2
## MPP7-DT MPP7-DT
## ATG9B.1 ATG9B
## LINC00539 LINC00539
## FAR1 FAR1
## NUP50-DT NUP50-DT
## ZBTB6 ZBTB6
## PHF7.1 PHF7
## AL133268.4 AL133268.4
## TRAV12-2.2 TRAV12-2
## PRKD3 PRKD3
## SLAMF7.1 SLAMF7
## ZNHIT6.1 ZNHIT6
## IPO13.1 IPO13
## Z94721.1.2 Z94721.1
## A1BG-AS1 A1BG-AS1
## HSDL1 HSDL1
## FOXN2 FOXN2
## ZSCAN16 ZSCAN16
## PEX14 PEX14
## CTSF CTSF
## SEC24D SEC24D
## APP.2 APP
## SYNJ2BP SYNJ2BP
## KLHL42.2 KLHL42
## MSTO1.1 MSTO1
## LYL1.1 LYL1
## CHTF18.2 CHTF18
## BICD2 BICD2
## GABPB1.1 GABPB1
## MATR3.1 MATR3.1
## NOP14-AS1 NOP14-AS1
## ZC3H18.1 ZC3H18
## CARD11.1 CARD11
## AC025171.1 AC025171.1
## LTO1 LTO1
## GATAD1 GATAD1
## ALMS1.1 ALMS1
## DHCR24.2 DHCR24
## TMEM237.2 TMEM237
## NFKBIE.1 NFKBIE
## STK40.1 STK40
## SELENON.2 SELENON
## FBXO25 FBXO25
## FAM160B2 FAM160B2
## CD248.1 CD248
## RMND5A.1 RMND5A
## PLEKHM2.1 PLEKHM2
## SECISBP2 SECISBP2
## CSGALNACT2 CSGALNACT2
## LMNA.1 LMNA
## TRBV19 TRBV19
## TWSG1 TWSG1
## ARHGEF39.2 ARHGEF39
## ZRSR2 ZRSR2
## ZBTB11 ZBTB11
## PHF12.1 PHF12
## XRCC3.2 XRCC3
## ALOX12-AS1 ALOX12-AS1
## KBTBD3 KBTBD3
## AL390066.1 AL390066.1
## EPSTI1.2 EPSTI1
## STRN4 STRN4
## KIFAP3 KIFAP3
## INTS2 INTS2
## CRNDE.2 CRNDE
## STAT6 STAT6
## ODF2L.1 ODF2L
## PDPK1 PDPK1
## ZFP62 ZFP62
## LZTS2 LZTS2
## CENPJ.2 CENPJ
## NEK1.1 NEK1
## APOBEC3D APOBEC3D
## PNPLA8 PNPLA8
## VPS35L VPS35L
## UTP20.1 UTP20
## NBPF19.1 NBPF19
## METTL4 METTL4
## AKAP8L.1 AKAP8L
## PUDP PUDP
## LRRC37A3 LRRC37A3
## WDR92 WDR92
## ITGA4.2 ITGA4
## ZNF430.1 ZNF430
## CENPP.2 CENPP
## ACTR8 ACTR8
## COQ3 COQ3
## FHL1.1 FHL1
## CMSS1.2 CMSS1
## OSBPL11 OSBPL11
## NMT2.1 NMT2
## TRIM26 TRIM26
## TTC21B TTC21B
## RNF146 RNF146
## USP4 USP4
## MAPKAPK2.1 MAPKAPK2
## PER1.1 PER1
## SYNJ2 SYNJ2
## ZNF557.1 ZNF557
## ANGEL1 ANGEL1
## PGBD2 PGBD2
## THAP8 THAP8
## TMX3 TMX3
## APOL6.1 APOL6
## RNF40 RNF40
## METTL2B METTL2B
## AC067945.4 AC067945.4
## TRIP4 TRIP4
## NFS1.1 NFS1
## AP3S2 AP3S2
## RCBTB1.1 RCBTB1
## PICK1 PICK1
## RARG RARG
## MFAP3 MFAP3
## DNAJA4.1 DNAJA4
## FAIM FAIM
## CDK6.1 CDK6
## BPHL.1 BPHL
## FURIN.1 FURIN
## REV1.2 REV1
## TPD52.1 TPD52
## SPAG1 SPAG1
## ZNF518B.1 ZNF518B
## MPHOSPH9.1 MPHOSPH9
## ASTE1 ASTE1
## EDEM3 EDEM3
## PARP12 PARP12
## AC006369.1.2 AC006369.1
## CCDC150.2 CCDC150
## TRIM56 TRIM56
## CYTH1 CYTH1
## SLC25A42 SLC25A42
## AL360178.1 AL360178.1
## TRMT12 TRMT12
## ALDH1B1 ALDH1B1
## CD200R1.1 CD200R1
## IRF5 IRF5
## NFATC3 NFATC3
## HIST1H2AH.2 HIST1H2AH
## SLC35E3 SLC35E3
## TRAV8-3 TRAV8-3
## HERC5 HERC5
## JADE1.1 JADE1
## GSTM3.1 GSTM3
## IP6K1 IP6K1
## PPP3CB-AS1 PPP3CB-AS1
## STAG3.2 STAG3
## NEK4 NEK4
## FLCN FLCN
## IDH1 IDH1
## VANGL1.1 VANGL1
## HLA-G HLA-G
## AQR.1 AQR
## PPFIA1 PPFIA1
## ZNF512 ZNF512
## THUMPD2.1 THUMPD2
## ZNF726.1 ZNF726
## TRAV39 TRAV39
## PET117 PET117
## ZYG11B.1 ZYG11B
## ZNF140 ZNF140
## SMIM8 SMIM8
## ZNF684.1 ZNF684
## METTL16.1 METTL16
## CPT2 CPT2
## DNM1L.1 DNM1L
## RRAS2.2 RRAS2
## FAM161A.2 FAM161A
## CC2D1A.1 CC2D1A
## SCAP SCAP
## BRD9.1 BRD9
## USP15 USP15
## GNL3.2 GNL3
## ORC2.1 ORC2
## AMACR AMACR
## FKBPL.1 FKBPL
## PRMT6 PRMT6
## LUC7L LUC7L
## MIOS MIOS
## KYAT1 KYAT1
## SGCB SGCB
## AL356356.1 AL356356.1
## ABHD5 ABHD5
## ZNF260 ZNF260
## STAT2.1 STAT2
## BRMS1L BRMS1L
## TTPAL TTPAL
## ZNF232.2 ZNF232
## TRAV13-1.1 TRAV13-1
## TRBV27.1 TRBV27
## ZNF599 ZNF599
## GFM2.1 GFM2
## ZW10.1 ZW10
## BIRC3.2 BIRC3
## CD6 CD6
## ICK ICK
## SLC30A7 SLC30A7
## ZNF691 ZNF691
## ZNF395 ZNF395
## UBXN8.1 UBXN8
## SDCCAG8 SDCCAG8
## MSL1 MSL1
## SLC2A11 SLC2A11
## TUBB4A.1 TUBB4A
## LOXL1-AS1.1 LOXL1-AS1
## MACROD1.2 MACROD1
## SIDT2 SIDT2
## ACOX3.1 ACOX3
## RLIM RLIM
## WAC WAC
## TRAV41 TRAV41
## CORO2A CORO2A
## SPINT1.1 SPINT1
## GAA GAA
## CACUL1 CACUL1
## NAA40.1 NAA40
## POLR3F POLR3F
## TXLNA.1 TXLNA
## ZDHHC14.2 ZDHHC14
## PANX1 PANX1
## GPR160.1 GPR160
## ABHD3 ABHD3
## TRAPPC9.1 TRAPPC9
## ACBD4 ACBD4
## ZNF107.2 ZNF107
## PPP2CB PPP2CB
## SIRT5.1 SIRT5
## SLC11A2.1 SLC11A2
## ASB2.1 ASB2
## AC005944.1 AC005944.1
## ERCC6L.2 ERCC6L
## PLCB2.1 PLCB2
## AL392172.1 AL392172.1
## PGAP3 PGAP3
## TRIM34 TRIM34
## FBXL8.1 FBXL8
## ROCK2.1 ROCK2
## RGP1 RGP1
## SCMH1.2 SCMH1
## SPATA33.1 SPATA33
## AKNA.1 AKNA
## ZNF200 ZNF200
## ZNF211 ZNF211
## USP19 USP19
## MAF.2 MAF
## SLC27A4 SLC27A4
## NRDC.1 NRDC
## FBXW9 FBXW9
## EFCAB2.1 EFCAB2
## KLHDC10 KLHDC10
## ESR2 ESR2
## ELF4 ELF4
## SOCS4.1 SOCS4
## LEMD2 LEMD2
## PAN2 PAN2
## KMT2B KMT2B
## RTN4IP1 RTN4IP1
## PCM1 PCM1
## ZNF701 ZNF701
## TBX21.2 TBX21
## FAM153CP FAM153CP
## COA7.1 COA7
## CCNG2.1 CCNG2
## ZNF275 ZNF275
## STRIP1 STRIP1
## SLC35F2.1 SLC35F2
## C9orf72.1 C9orf72
## PCTP PCTP
## FAF1.1 FAF1
## SPIN1.1 SPIN1
## TRAV22 TRAV22
## COPG2.2 COPG2
## ARHGAP33.2 ARHGAP33
## ZNF286A ZNF286A
## ZBTB22 ZBTB22
## THAP2 THAP2
## ERLIN2.1 ERLIN2
## FOXP1.1 FOXP1
## CLSTN3.1 CLSTN3
## TRAV25 TRAV25
## RAB33B RAB33B
## SLA SLA
## DLG3 DLG3
## HDAC7.1 HDAC7
## TEFM TEFM
## GORAB GORAB
## POMC POMC
## ABCC10 ABCC10
## FBXO21 FBXO21
## INF2.1 INF2
## ANKRD49 ANKRD49
## RRNAD1 RRNAD1
## PEX1 PEX1
## KMT5C KMT5C
## CCDC106.1 CCDC106
## ARSK.1 ARSK
## ZNF181 ZNF181
## TRAV36DV7 TRAV36DV7
## MFSD14B.1 MFSD14B
## ZDHHC13.1 ZDHHC13
## RCOR3.1 RCOR3
## HPGD.1 HPGD
## MOV10.2 MOV10
## DNAJB2 DNAJB2
## CRACR2B.1 CRACR2B
## TOB2 TOB2
## AK5.1 AK5
## FASN.2 FASN
## URGCP.1 URGCP
## DZIP3.1 DZIP3
## SPATA5L1 SPATA5L1
## CPNE7 CPNE7
## SLC16A1.2 SLC16A1
## GLYCTK GLYCTK
## PAOX.1 PAOX
## CXorf56 CXorf56
## ELF2 ELF2
## ZAP70.2 ZAP70
## WDR24 WDR24
## PAFAH1B1 PAFAH1B1
## PPP3CB PPP3CB
## STK3.2 STK3
## TRIM32 TRIM32
## RASA3.1 RASA3
## ZNF740 ZNF740
## NCAM1.2 NCAM1
## ATAD3B.1 ATAD3B
## CENPBD1 CENPBD1
## CEP68 CEP68
## ZNF724.1 ZNF724
## FAM86C1 FAM86C1
## SIAH1.1 SIAH1
## SUGP2.1 SUGP2
## CTSO CTSO
## YTHDC2.1 YTHDC2
## AC012447.1.1 AC012447.1
## SENP2 SENP2
## AC147067.1.1 AC147067.1
## GSK3B.1 GSK3B
## DCP1B DCP1B
## SC5D SC5D
## CAPN10 CAPN10
## CD247.1 CD247
## PLOD1.1 PLOD1
## AC006449.6 AC006449.6
## KIAA1328.2 KIAA1328
## DBF4B.2 DBF4B
## SNHG11 SNHG11
## HELLPAR.2 HELLPAR
## FXR2 FXR2
## VPS41 VPS41
## TRIM44.2 TRIM44
## USP34 USP34
## PRR11.2 PRR11
## GATB.1 GATB
## DYRK1B.1 DYRK1B
## AC008035.1.1 AC008035.1
## FAM199X FAM199X
## RARS2 RARS2
## ANKRD13C ANKRD13C
## UBL7-AS1.2 UBL7-AS1
## ARIH2OS ARIH2OS
## FANCC.2 FANCC
## LRMP LRMP
## NUMA1 NUMA1
## CCDC159 CCDC159
## LTBP3.1 LTBP3
## NSRP1 NSRP1
## ZBTB7B.2 ZBTB7B
## SETD6 SETD6
## TMEM175 TMEM175
## ZNF37A.1 ZNF37A
## FRAT1 FRAT1
## FBXO38 FBXO38
## ANKRD27 ANKRD27
## ZNF429 ZNF429
## LIN9.2 LIN9
## RHOU.1 RHOU
## MISP3 MISP3
## MOCS3 MOCS3
## HOTAIRM1.1 HOTAIRM1
## AP003717.4.1 AP003717.4
## NEFL.2 NEFL
## UIMC1 UIMC1
## CLUH.2 CLUH
## ZNF394.1 ZNF394
## LY75 LY75
## PACS2.1 PACS2
## KCTD2 KCTD2
## SBNO1-AS1 SBNO1-AS1
## CBWD2 CBWD2
## OPN3 OPN3
## PRSS21 PRSS21
## EDC3 EDC3
## AC053513.1 AC053513.1
## PPIP5K2.2 PPIP5K2
## GPR180.1 GPR180
## WDR35 WDR35
## ZSCAN26 ZSCAN26
## MAP3K20.2 MAP3K20
## ZNF792 ZNF792
## C1RL C1RL
## SEMA4C SEMA4C
## CLECL1.1 CLECL1
## HEATR1.1 HEATR1
## IL17RA IL17RA
## ZBTB10 ZBTB10
## TCTA.1 TCTA
## RPARP-AS1 RPARP-AS1
## QPCTL QPCTL
## MTOR MTOR
## KDM6A KDM6A
## FUBP1.1 FUBP1
## ZER1 ZER1
## MORC4.1 MORC4
## NR2C1 NR2C1
## MANEAL.1 MANEAL
## AC022217.3.1 AC022217.3
## AP006621.3 AP006621.3
## COL18A1.1 COL18A1
## PRKCZ PRKCZ
## C10orf143 C10orf143
## GNLY.1 GNLY
## CALHM6.2 CALHM6
## DLGAP1-AS1 DLGAP1-AS1
## SAV1.2 SAV1
## CLASP1.1 CLASP1
## P2RY14.2 P2RY14
## TRAV19 TRAV19
## EXOC8 EXOC8
## CSNK1G3.1 CSNK1G3
## RPP14 RPP14
## EDRF1.1 EDRF1
## LINC01934 LINC01934
## CSNK1E CSNK1E
## AC016074.2 AC016074.2
## RASAL3.1 RASAL3
## PKD2L2 PKD2L2
## DUSP7 DUSP7
## LIG3 LIG3
## BCL7A.1 BCL7A
## SYCE2.2 SYCE2
## CA11 CA11
## USP3-AS1 USP3-AS1
## OSBPL2.1 OSBPL2
## HECTD1 HECTD1
## ETS1.1 ETS1
## RBM27 RBM27
## C4orf33.1 C4orf33
## PRUNE1 PRUNE1
## SMARCAL1 SMARCAL1
## RDM1 RDM1
## HOOK3.1 HOOK3
## FANCA.2 FANCA
## WDR60 WDR60
## AGPS.2 AGPS
## ZSCAN25 ZSCAN25
## LYRM9 LYRM9
## MAML2.1 MAML2
## TJAP1 TJAP1
## SVIL.2 SVIL
## GGACT GGACT
## AC087854.1 AC087854.1
## TMC8 TMC8
## NCMAP-DT NCMAP-DT
## LINC01006 LINC01006
## RBMXL1 RBMXL1
## CARMIL2.1 CARMIL2
## CERS5.2 CERS5
## SAPCD2.2 SAPCD2
## AL022157.1 AL022157.1
## DHRS11 DHRS11
## GPATCH1.1 GPATCH1
## B3GLCT B3GLCT
## ZNF431.1 ZNF431
## H2AFY2 H2AFY2
## CTBP1-AS CTBP1-AS
## ARL13B.1 ARL13B
## ITGB2-AS1.1 ITGB2-AS1
## CTLA4.2 CTLA4
## C19orf47.1 C19orf47
## OSGEPL1 OSGEPL1
## MIR762HG MIR762HG
## ZNF780A ZNF780A
## ICE1 ICE1
## HPS3 HPS3
## SS18 SS18
## B3GNTL1.1 B3GNTL1
## SRSF7 SRSF7
## TRDMT1.1 TRDMT1
## NAA30 NAA30
## FAM117B.1 FAM117B
## GINS4.2 GINS4
## AC073896.2 AC073896.2
## ZNF506 ZNF506
## CCL5.2 CCL5
## SNHG21 SNHG21
## FAM13B FAM13B
## CTNNA1 CTNNA1
## CNTRL CNTRL
## RLF RLF
## SYT11.1 SYT11
## MDC1.1 MDC1
## ILF3-DT.1 ILF3-DT
## FAM76A FAM76A
## NAA10.1 NAA10
## RBM4B RBM4B
## CEP76.2 CEP76
## WDR53.1 WDR53
## CARM1.1 CARM1
## CYP4V2 CYP4V2
## WDSUB1.1 WDSUB1
## SACS.1 SACS
## B3GNT9 B3GNT9
## EVL.1 EVL
## TRAV14DV4.1 TRAV14DV4
## ATF6.1 ATF6
## PLEKHA2 PLEKHA2
## AC079921.1.1 AC079921.1
## LANCL2.1 LANCL2
## YPEL2.1 YPEL2
## ANAPC1.1 ANAPC1
## LRRC23.1 LRRC23
## AC009133.3.1 AC009133.3
## LIMS2.1 LIMS2
## FAM241B.2 FAM241B
## C5orf34.1 C5orf34
## AL096870.8 AL096870.8
## TIGD5 TIGD5
## CEP131.1 CEP131
## NUAK2.1 NUAK2
## LINC02482 LINC02482
## MAP3K2-DT MAP3K2-DT
## C5orf22 C5orf22
## CCNE2.2 CCNE2
## ADCY9.1 ADCY9
## GTF2A1 GTF2A1
## CENPI.2 CENPI
## ZNF143.1 ZNF143
## PNISR.2 PNISR
## ZNF675 ZNF675
## APPL2 APPL2
## RPS6KB1 RPS6KB1
## OXSM OXSM
## SLC4A7 SLC4A7
## FBXO46 FBXO46
## KPNA1 KPNA1
## CDR2 CDR2
## RCCD1.2 RCCD1
## ZC3H4.1 ZC3H4
## ZC3H12A ZC3H12A
## MFSD8 MFSD8
## TRAIP.2 TRAIP
## C14orf28 C14orf28
## POLR1B POLR1B
## ATP13A2 ATP13A2
## TRPC4AP.1 TRPC4AP
## NCOA5.1 NCOA5
## ETV7.2 ETV7
## FUT8 FUT8
## INTS4 INTS4
## PPP1R12A.1 PPP1R12A
## ZRANB3.2 ZRANB3
## ADAMTSL4-AS1.2 ADAMTSL4-AS1
## BAHD1 BAHD1
## MPRIP MPRIP
## RARA RARA
## CEP170.1 CEP170
## DCLRE1B.2 DCLRE1B
## AC021028.1 AC021028.1
## SMCHD1 SMCHD1
## CHD1L.1 CHD1L
## LAX1.1 LAX1
## DUSP18 DUSP18
## NUFIP2.1 NUFIP2
## CHROMR CHROMR
## URB1-AS1 URB1-AS1
## DCLRE1C.1 DCLRE1C
## CEP95.1 CEP95
## LINC00624 LINC00624
## DNAJC13 DNAJC13
## LRRC8A LRRC8A
## WDR45B WDR45B
## LINC02762 LINC02762
## ABHD6 ABHD6
## NSUN4 NSUN4
## PAQR8.1 PAQR8
## ALG9.1 ALG9
## MARCH3.2 MARCH3
## SLC39A9.1 SLC39A9
## GPR174.2 GPR174
## MBTPS2 MBTPS2
## AC004865.2.1 AC004865.2
## DCAF17 DCAF17
## MAP3K3 MAP3K3
## MORN3 MORN3
## BTLA BTLA
## EML2-AS1 EML2-AS1
## ARRB1.1 ARRB1
## AL590764.1 AL590764.1
## ZBTB45 ZBTB45
## FAM89A FAM89A
## SPATA24 SPATA24
## SOX12 SOX12
## PSKH1 PSKH1
## TPM1.1 TPM1
## IL23R.2 IL23R
## TMEM150A.1 TMEM150A
## S100PBP S100PBP
## METTL8.2 METTL8
## TSPAN15 TSPAN15
## HCFC2 HCFC2
## SNX8.2 SNX8
## TST TST
## SCRN1 SCRN1
## ASNSD1.1 ASNSD1
## KLRG1.2 KLRG1
## BCOR.1 BCOR
## TMEM116 TMEM116
## FUT11 FUT11
## CC2D1B CC2D1B
## KLHL36 KLHL36
## RIPOR1 RIPOR1
## ARNT ARNT
## TRAF3IP1.1 TRAF3IP1
## AKAP10.1 AKAP10
## ECD ECD
## TMEM91 TMEM91
## METAP1D.1 METAP1D
## ZNF718.1 ZNF718
## DCAF16.1 DCAF16
## HPDL.1 HPDL
## STX18.1 STX18
## AC007114.1 AC007114.1
## VPS33B.1 VPS33B
## KNTC1.2 KNTC1
## ZNF419 ZNF419
## NADSYN1 NADSYN1
## RFESD.1 RFESD
## KIF3B KIF3B
## ZNF451 ZNF451
## GPR15.1 GPR15
## FILIP1L.1 FILIP1L
## ZNF213.1 ZNF213
## OPA1 OPA1
## ZNF561 ZNF561
## PJA2 PJA2
## TTL TTL
## ERN1.1 ERN1
## AC007384.1.1 AC007384.1
## PLEKHO2 PLEKHO2
## NF2.2 NF2
## C20orf96.1 C20orf96
## CATSPERB CATSPERB
## ATP1B1.2 ATP1B1
## CEP89.1 CEP89
## ZNF8 ZNF8
## CERS6.2 CERS6
## MPP7.1 MPP7
## SULT1B1 SULT1B1
## ELP3 ELP3
## HACD2 HACD2
## REL.1 REL
## PIMREG.2 PIMREG
## AL627171.1 AL627171.1
## PARN.1 PARN
## ORC1.2 ORC1
## KLRC2.2 KLRC2
## TRAV38-2DV8 TRAV38-2DV8
## FCHSD1.1 FCHSD1
## ZNF134 ZNF134
## MICAL1.1 MICAL1
## ANKMY2 ANKMY2
## DUS4L DUS4L
## PPP4R3B PPP4R3B
## ATL2.1 ATL2
## GNE GNE
## G2E3.1 G2E3
## ZNF761 ZNF761
## TRGV5.1 TRGV5
## CCDC88A.2 CCDC88A
## AL365361.1 AL365361.1
## STX11.1 STX11
## OGFRL1 OGFRL1
## RTKN2.2 RTKN2
## CASD1 CASD1
## REC8.1 REC8
## GIGYF1 GIGYF1
## FAM122C FAM122C
## TBC1D23 TBC1D23
## KHDC4.1 KHDC4
## KAT7 KAT7
## AC044849.1 AC044849.1
## FOXJ2 FOXJ2
## THTPA THTPA
## ZFX ZFX
## ATXN7L2 ATXN7L2
## PYCR3 PYCR3
## APOM APOM
## INPP5E INPP5E
## CTBP1-DT.1 CTBP1-DT
## LYSMD3 LYSMD3
## CEP350 CEP350
## MIR155HG.1 MIR155HG
## FAM168B FAM168B
## ZNF184 ZNF184
## SLC4A2 SLC4A2
## ELOVL4.1 ELOVL4
## RAB3IP.2 RAB3IP
## DEPDC1.2 DEPDC1
## USP45 USP45
## USP7 USP7
## NLRP2 NLRP2
## AL136419.3.1 AL136419.3
## DPYSL2.1 DPYSL2
## ANTXR2.1 ANTXR2
## HDAC6 HDAC6
## SLC26A6.1 SLC26A6
## CDKAL1.1 CDKAL1
## SLC25A15 SLC25A15
## CYP2R1 CYP2R1
## ZNF23.1 ZNF23
## AC139530.1 AC139530.1
## CREB3L4 CREB3L4
## RAC3.2 RAC3
## FAM102B.2 FAM102B
## MIEF2 MIEF2
## ZNF473.2 ZNF473
## PARP14.2 PARP14
## SLC25A16.1 SLC25A16
## MMEL1.1 MMEL1
## AL139246.5.1 AL139246.5
## ATP1A1-AS1 ATP1A1-AS1
## ALKBH1 ALKBH1
## RPAP2 RPAP2
## FYN.2 FYN
## FAM160A2.1 FAM160A2
## GALNT6.1 GALNT6
## PFKM.1 PFKM
## ZNF717 ZNF717
## CNBD2 CNBD2
## SDE2 SDE2
## DENND11 DENND11
## CHRM3 CHRM3
## EIF4ENIF1 EIF4ENIF1
## DNAL1 DNAL1
## AIFM2 AIFM2
## ARNTL2.2 ARNTL2
## CCZ1B CCZ1B
## ZNF331.1 ZNF331
## PHF20 PHF20
## MSH5.1 MSH5
## AC006504.5 AC006504.5
## DPY19L1.1 DPY19L1
## KHDC1.1 KHDC1
## AC025159.1 AC025159.1
## FAM193A.1 FAM193A
## PREP PREP
## NUDT19 NUDT19
## SKIV2L SKIV2L
## GPR89A GPR89A
## GALM.1 GALM
## NCK1 NCK1
## C14orf93 C14orf93
## TMEM30A.1 TMEM30A
## RMND5B.2 RMND5B
## ALG14.1 ALG14
## RBM41 RBM41
## UHRF1BP1L UHRF1BP1L
## ATG13.1 ATG13
## FBXW7.1 FBXW7
## NR1H3 NR1H3
## TRBV7-6.1 TRBV7-6
## AC090152.1.2 AC090152.1
## ATG9A ATG9A
## SLC7A6 SLC7A6
## SPATA2 SPATA2
## CCDC50 CCDC50
## TM6SF1.1 TM6SF1
## RAB28.1 RAB28
## KMO KMO
## ZNF92 ZNF92
## PIP4K2B PIP4K2B
## CCL4L2.2 CCL4L2
## TRAF5.1 TRAF5
## AGFG1.1 AGFG1
## GATD1 GATD1
## MDM2 MDM2
## PROSER1 PROSER1
## FOSL2.1 FOSL2
## DHODH DHODH
## ZNRF1.1 ZNRF1
## ERLIN1.1 ERLIN1
## CHKA CHKA
## AL606489.1 AL606489.1
## ATP13A3.1 ATP13A3
## HAUS5.2 HAUS5
## HIST1H2AB.1 HIST1H2AB
## NOL10.1 NOL10
## ZNF821 ZNF821
## EPOR EPOR
## NCF2 NCF2
## LINC00526 LINC00526
## MLH3 MLH3
## AC211476.2 AC211476.2
## FKBP1C FKBP1C
## CMIP CMIP
## PDE5A PDE5A
## HILPDA.1 HILPDA
## MPP6.1 MPP6
## CPNE2 CPNE2
## XYLB.2 XYLB
## KIAA0232.1 KIAA0232
## UTP25 UTP25
## BRPF1 BRPF1
## MTMR1 MTMR1
## SHLD2.1 SHLD2
## ZNF699 ZNF699
## NAP1L5 NAP1L5
## DHX32 DHX32
## AC026979.2 AC026979.2
## COQ7 COQ7
## GSTCD.2 GSTCD
## MPHOSPH8 MPHOSPH8
## AC090204.1.1 AC090204.1
## RNF185 RNF185
## NSMAF.1 NSMAF
## CNOT6 CNOT6
## TMED8.1 TMED8
## BIVM BIVM
## WRN.2 WRN
## TRBV5-1 TRBV5-1
## GIN1 GIN1
## MLKL.1 MLKL
## CCDC84-DT.2 CCDC84-DT
## ZNF182 ZNF182
## BEX5 BEX5
## SPEF2 SPEF2
## CCDC200.1 CCDC200
## AC021683.5 AC021683.5
## ZNF513 ZNF513
## HDAC5.1 HDAC5
## DUSP4 DUSP4
## TSHZ2.2 TSHZ2
## NIN.1 NIN
## AMD1 AMD1
## HNRNPLL.2 HNRNPLL
## PRPF4B PRPF4B
## BOLA2B BOLA2B
## PTDSS2.1 PTDSS2
## CNGA4 CNGA4
## SLC7A1.2 SLC7A1
## ZXDC ZXDC
## SENP1.1 SENP1
## RAB43.1 RAB43
## RC3H2.1 RC3H2
## APOBEC3B.2 APOBEC3B
## ABHD15-AS1 ABHD15-AS1
## PDPR PDPR
## SORD SORD
## AC018521.5 AC018521.5
## ZNF345 ZNF345
## AP002490.1 AP002490.1
## ZBP1.1 ZBP1
## ACTL10 ACTL10
## PCBD2.2 PCBD2
## FAM214B FAM214B
## KMT5B KMT5B
## TRBJ2-3 TRBJ2-3
## LDAH LDAH
## PIGM PIGM
## MYO19.2 MYO19
## SPG7.1 SPG7
## ZNF681.1 ZNF681
## RINT1 RINT1
## PPP2R5B PPP2R5B
## PTPRA PTPRA
## UBE2H UBE2H
## ZNF846 ZNF846
## NOP9 NOP9
## C2orf27A C2orf27A
## NRAV NRAV
## SCAF4.1 SCAF4
## SWAP70.1 SWAP70
## SLC25A29 SLC25A29
## LARP1B.2 LARP1B
## MAPK11 MAPK11
## TLE3 TLE3
## ACAP1.1 ACAP1
## SYNGAP1 SYNGAP1
## COG7 COG7
## HMG20A.1 HMG20A
## MBTPS1 MBTPS1
## KLHL21 KLHL21
## CLIP1.1 CLIP1
## PRMT3.2 PRMT3
## ZNF17 ZNF17
## ZNF398 ZNF398
## GALNT12.2 GALNT12
## PKD2.1 PKD2
## AC245407.2.1 AC245407.2
## UBE3B.1 UBE3B
## FAM160B1 FAM160B1
## ZBTB9 ZBTB9
## GFOD1.2 GFOD1
## HIRA HIRA
## NMNAT1 NMNAT1
## CHST7.1 CHST7
## BFSP2.1 BFSP2
## TOGARAM1 TOGARAM1
## ABCD1.1 ABCD1
## ZNF575 ZNF575
## AC100861.1 AC100861.1
## SPHK1 SPHK1
## SMTN.1 SMTN
## FAM72B.2 FAM72B
## CCL4.2 CCL4
## ZSCAN2 ZSCAN2
## SENCR SENCR
## TAPT1.1 TAPT1
## KBTBD7 KBTBD7
## NRGN.2 NRGN
## AC004556.3.1 AC004556.3
## TCAF1.1 TCAF1
## SESN2.1 SESN2
## ECPAS ECPAS
## USP18.1 USP18
## KLF11.1 KLF11
## SP140L.1 SP140L
## DPY19L4 DPY19L4
## C5orf30.1 C5orf30
## SUSD6.1 SUSD6
## MEF2D.1 MEF2D
## RETSAT RETSAT
## ENTPD1-AS1.1 ENTPD1-AS1
## P3H1 P3H1
## VIPR1.1 VIPR1
## DYNC2LI1 DYNC2LI1
## DFFB DFFB
## VAC14.1 VAC14
## INO80D INO80D
## ZNF507 ZNF507
## CREBRF CREBRF
## ANKRD10 ANKRD10
## ALKBH8 ALKBH8
## SP1.1 SP1
## NCK1-DT NCK1-DT
## ASRGL1.2 ASRGL1
## DHDDS.1 DHDDS
## ATG4A ATG4A
## SLC25A35 SLC25A35
## BCL2L11.1 BCL2L11
## LACTB2-AS1 LACTB2-AS1
## PTPRO.1 PTPRO
## RAB11B-AS1 RAB11B-AS1
## NADK2 NADK2
## CAD.1 CAD
## E2F7.2 E2F7
## DPH6.2 DPH6
## STAM2 STAM2
## LINC01393 LINC01393
## CARNMT1.1 CARNMT1
## PUS7.2 PUS7
## C16orf95.2 C16orf95
## ARHGEF19 ARHGEF19
## ENTPD5.1 ENTPD5
## ROPN1L ROPN1L
## C17orf53.2 C17orf53
## KDM2B KDM2B
## CROT CROT
## ABTB2.1 ABTB2
## CLIC5.2 CLIC5
## CKAP4.1 CKAP4
## ATF2 ATF2
## RNF170 RNF170
## OSGIN2 OSGIN2
## CENPC CENPC
## ZKSCAN8 ZKSCAN8
## AC104365.1 AC104365.1
## AUNIP.2 AUNIP
## C2orf69 C2orf69
## MUC1 MUC1
## SDC4.1 SDC4
## ETV2 ETV2
## NFIL3 NFIL3
## AC099063.1.1 AC099063.1
## AC005837.1 AC005837.1
## NT5M NT5M
## ACSS2.2 ACSS2
## NSUN3 NSUN3
## SLC35B4 SLC35B4
## ZNF273.2 ZNF273
## AC103563.7 AC103563.7
## WDCP WDCP
## PTER PTER
## TMEM201.2 TMEM201
## PIGB PIGB
## PILRA PILRA
## CPPED1.1 CPPED1
## SBF1 SBF1
## PLCL1.2 PLCL1
## SPOPL.1 SPOPL
## AGPAT4.2 AGPAT4
## TAB1 TAB1
## PRXL2A.2 PRXL2A
## TRBV11-2 TRBV11-2
## ZNF621 ZNF621
## IFNL1.1 IFNL1
## BFSP1.1 BFSP1
## INSL6 INSL6
## TECPR1 TECPR1
## EPS15L1 EPS15L1
## IVD.1 IVD
## ANKRD16.1 ANKRD16
## SDHAF4 SDHAF4
## PBRM1.1 PBRM1
## PITRM1.1 PITRM1
## NEK2.2 NEK2
## EXO5 EXO5
## DDI2 DDI2
## ZMAT3.1 ZMAT3
## HIC2.1 HIC2
## CCDC126 CCDC126
## ANKAR ANKAR
## ACER3.1 ACER3
## RUNX2.2 RUNX2
## ERCC6L2 ERCC6L2
## ZNF525 ZNF525
## SDCBP2-AS1 SDCBP2-AS1
## NOL9 NOL9
## ATG14.1 ATG14
## NHLRC4 NHLRC4
## AC008440.1 AC008440.1
## METTL14 METTL14
## FAM13A.1 FAM13A
## SHPK.1 SHPK
## PTPN1.1 PTPN1
## ZSCAN29 ZSCAN29
## IDI1.1 IDI1
## PDS5A PDS5A
## RNF123 RNF123
## SYCE1L SYCE1L
## SCAMP1 SCAMP1
## PGGT1B.1 PGGT1B
## KMT2E-AS1 KMT2E-AS1
## FBXO31 FBXO31
## DGCR2 DGCR2
## PARP16.2 PARP16
## ARMC7 ARMC7
## LZTR1 LZTR1
## TBC1D31.1 TBC1D31
## KIAA0100.1 KIAA0100
## PPP1R3D PPP1R3D
## HIPK3 HIPK3
## RPAP1 RPAP1
## HAVCR2.1 HAVCR2
## SLC25A53 SLC25A53
## ETS2 ETS2
## ASH1L-AS1 ASH1L-AS1
## ZNF133 ZNF133
## PCNX4 PCNX4
## TRIM41.1 TRIM41
## CTNNB1 CTNNB1
## SLC37A3 SLC37A3
## TP53I3 TP53I3
## ZNF530.1 ZNF530
## DCAKD.1 DCAKD
## SH2D3C.1 SH2D3C
## PCSK1N.1 PCSK1N
## AK1 AK1
## AL137857.1 AL137857.1
## MTREX MTREX
## CLN8 CLN8
## WARS2 WARS2
## MGRN1 MGRN1
## MAPK6.1 MAPK6
## IPO11.1 IPO11
## AL080276.2 AL080276.2
## ZKSCAN5 ZKSCAN5
## SLC35E1.1 SLC35E1
## TMEM143 TMEM143
## HOXB-AS1 HOXB-AS1
## SIL1.1 SIL1
## ZNF383 ZNF383
## PDLIM5.1 PDLIM5
## PRKAB2 PRKAB2
## URB2 URB2
## DMWD DMWD
## PRKX PRKX
## UPF2.1 UPF2
## CCR5.2 CCR5
## CLIP3 CLIP3
## SLC49A4 SLC49A4
## AC023509.6 AC023509.6
## AC022182.2.1 AC022182.2
## MICU1.1 MICU1
## MTPAP.1 MTPAP
## CDCP1 CDCP1
## CACTIN CACTIN
## ZNF189 ZNF189
## AL451165.2 AL451165.2
## RECK.1 RECK
## PRMT9.1 PRMT9
## HELQ HELQ
## RGL2.1 RGL2
## NT5DC2.1 NT5DC2
## NUDCD3.1 NUDCD3
## RDH13 RDH13
## ZNF493 ZNF493
## NDUFAF5 NDUFAF5
## AC108488.1 AC108488.1
## SNURF SNURF
## ENOX2 ENOX2
## MARS2 MARS2
## TRAV18 TRAV18
## SASH1 SASH1
## AC069224.1 AC069224.1
## BRAP BRAP
## ZBTB44 ZBTB44
## ZNF136 ZNF136
## VARS2.1 VARS2
## PIGG PIGG
## ACAP3 ACAP3
## RNF216.1 RNF216
## WDR81 WDR81
## AC131571.1 AC131571.1
## ANKUB1 ANKUB1
## PTBP2.1 PTBP2
## MTRF1 MTRF1
## ARHGAP12 ARHGAP12
## ARFGAP1 ARFGAP1
## RRBP1 RRBP1
## TRBV20-1 TRBV20-1
## ABI1.1 ABI1
## DOK6.1 DOK6
## TRAV21 TRAV21
## PLCXD1 PLCXD1
## AC008063.1.1 AC008063.1
## MIER3 MIER3
## LPAR5.1 LPAR5
## MAP4K3-DT MAP4K3-DT
## CDKL3.1 CDKL3
## ZNF486 ZNF486
## TEX10 TEX10
## PELI3 PELI3
## LY6E-DT LY6E-DT
## CHML CHML
## ZNF485 ZNF485
## ARL11 ARL11
## TLCD3A TLCD3A
## CCDC77.1 CCDC77
## AGO1 AGO1
## AP1G1.1 AP1G1
## NDOR1.1 NDOR1
## MTHFSD.1 MTHFSD
## NUS1 NUS1
## ZNF605 ZNF605
## MED22 MED22
## CCNL2 CCNL2
## KLHL8.1 KLHL8
## DENND1B.1 DENND1B
## TRAV26-2.1 TRAV26-2
## SLF2.1 SLF2
## PPP2R2A.1 PPP2R2A
## ZNF141.1 ZNF141
## NUP188.2 NUP188
## TRBV4-2.1 TRBV4-2
## MIER2.1 MIER2
## CLTC CLTC
## ZNF845 ZNF845
## PEX13 PEX13
## CLCN7 CLCN7
## EHD4 EHD4
## C3orf33 C3orf33
## AP003307.1 AP003307.1
## GPR150 GPR150
## ARL17B.1 ARL17B
## AC019163.1 AC019163.1
## MINCR MINCR
## TIGD1 TIGD1
## EMSY EMSY
## RAB2B RAB2B
## ZNF142 ZNF142
## IL2RA.1 IL2RA
## KDM5D.1 KDM5D
## KANSL1.1 KANSL1
## PAXIP1.2 PAXIP1
## EXT1.2 EXT1
## WDR3 WDR3
## ATXN7L3 ATXN7L3
## USP12 USP12
## FAM229B FAM229B
## DHFR2 DHFR2
## MYCBP2-AS1 MYCBP2-AS1
## TSPAN5.1 TSPAN5
## TMEM161B.1 TMEM161B
## TANK TANK
## ANKRD35.1 ANKRD35
## EID2B EID2B
## GTF2IRD2B GTF2IRD2B
## GAPVD1 GAPVD1
## ETAA1 ETAA1
## FADS1 FADS1
## ZNF57 ZNF57
## ZNRF2 ZNRF2
## MRPL30 MRPL30
## RUNX3.1 RUNX3
## AC104162.2.1 AC104162.2
## ZNF814 ZNF814
## RALGDS.1 RALGDS
## NCKIPSD.1 NCKIPSD
## TMEM234 TMEM234
## ARMC8 ARMC8
## ACO1 ACO1
## MED29 MED29
## LONP2 LONP2
## SGPP1.1 SGPP1
## APBB3 APBB3
## SMARCAD1.1 SMARCAD1
## GCA.2 GCA
## MICA MICA
## PIP5K1C PIP5K1C
## LINC02256.1 LINC02256
## ATP8A1.1 ATP8A1
## TRAV20.1 TRAV20
## BRF1.2 BRF1
## HECW2-AS1.1 HECW2-AS1
## AP001816.1 AP001816.1
## STX12 STX12
## KLRC1.2 KLRC1
## ASB13.1 ASB13
## LMF1 LMF1
## FAM83D.2 FAM83D
## CCDC65.1 CCDC65
## RBKS RBKS
## AL162258.2 AL162258.2
## KDM5A.1 KDM5A
## TAF13 TAF13
## MED23 MED23
## LIN54.2 LIN54
## TSPAN18.1 TSPAN18
## ZCCHC2.1 ZCCHC2
## ZFP1 ZFP1
## SLAMF1.1 SLAMF1
## F8 F8
## POLR3D POLR3D
## CCSAP.1 CCSAP
## TRRAP.1 TRRAP
## F2R.1 F2R
## SLC39A14.1 SLC39A14
## MPZL1.1 MPZL1
## TMEM245 TMEM245
## TSEN2 TSEN2
## ZNF93.2 ZNF93
## MIA2.2 MIA2
## ZC3H7B ZC3H7B
## REXO5 REXO5
## JARID2.1 JARID2
## BDP1.1 BDP1
## C1orf56.1 C1orf56
## ADAL.1 ADAL
## TCFL5 TCFL5
## TMEM156.1 TMEM156
## AC105446.1 AC105446.1
## EXTL2 EXTL2
## AL161421.1 AL161421.1
## AL109628.2 AL109628.2
## AACS.1 AACS
## SLC25A37.2 SLC25A37
## CEPT1.1 CEPT1
## MARCH7 MARCH7
## ABCD2.1 ABCD2
## HS2ST1.1 HS2ST1
## TTC27.1 TTC27
## ARHGAP21.1 ARHGAP21
## EOMES.1 EOMES
## SETBP1.2 SETBP1
## EIF3J-DT EIF3J-DT
## GTF2H2 GTF2H2
## KIF20A.2 KIF20A
## MPP5.1 MPP5
## CRK.1 CRK
## ATP10A.1 ATP10A
## ATG10.1 ATG10
## AKAP11 AKAP11
## TSTD2 TSTD2
## CKB CKB
## VPS39 VPS39
## TMEM104 TMEM104
## DROSHA.1 DROSHA
## KIAA0930 KIAA0930
## HBEGF HBEGF
## SBF2-AS1.1 SBF2-AS1
## SLC22A5.1 SLC22A5
## PWWP2B PWWP2B
## TMEM191C TMEM191C
## ENTPD3-AS1.1 ENTPD3-AS1
## XRN1.1 XRN1
## ZNF526 ZNF526
## CYTH4.1 CYTH4
## FAM49B.1 FAM49B
## GXYLT1.2 GXYLT1
## ZNF268 ZNF268
## TRMT2B.1 TRMT2B
## CASC1 CASC1
## CLPB.1 CLPB
## CDC73.1 CDC73
## PLCG1 PLCG1
## CDK5RAP2.2 CDK5RAP2
## CSKMT.1 CSKMT
## BTBD10 BTBD10
## TMEM161B-AS1 TMEM161B-AS1
## N4BP2.1 N4BP2
## RELT.1 RELT
## RB1CC1.1 RB1CC1
## ARL6 ARL6
## AC096677.1 AC096677.1
## AC253572.2.1 AC253572.2
## TMEM67 TMEM67
## ZEB1-AS1.1 ZEB1-AS1
## AC008443.1 AC008443.1
## TBCEL TBCEL
## SNX19 SNX19
## FPGT.1 FPGT
## ADCY3.1 ADCY3
## POLE.2 POLE
## ANKEF1 ANKEF1
## ZNF654.1 ZNF654
## ZNF280C ZNF280C
## HEXIM2 HEXIM2
## DGCR8.1 DGCR8
## AC002310.1 AC002310.1
## S100A5 S100A5
## IQGAP3.2 IQGAP3
## PRKAG2-AS1.1 PRKAG2-AS1
## TUBGCP5 TUBGCP5
## KDM3A KDM3A
## DNAJC27 DNAJC27
## DHX37 DHX37
## UPF1.1 UPF1
## PRKD2 PRKD2
## LINC01176 LINC01176
## TPRG1L TPRG1L
## YPEL1.1 YPEL1
## POLK.1 POLK
## PI4K2B.1 PI4K2B
## IGFBP4.2 IGFBP4
## TRBV6-5.1 TRBV6-5
## FAM241A.2 FAM241A
## MYB.2 MYB
## SHLD3 SHLD3
## CRTAM.2 CRTAM
## SH3BP5-AS1 SH3BP5-AS1
## ZNF777 ZNF777
## HMGCR HMGCR
## AC008083.3.1 AC008083.3
## SLC25A4 SLC25A4
## HIST1H2BD.2 HIST1H2BD
## DDIT4-AS1.1 DDIT4-AS1
## ZNF746 ZNF746
## AC007383.2 AC007383.2
## SERGEF SERGEF
## AC097382.3 AC097382.3
## CRIP2.1 CRIP2
## SBK1.1 SBK1
## KLHL24.1 KLHL24
## ZNF529 ZNF529
## MTMR9 MTMR9
## IL18R1.1 IL18R1
## CLASP2 CLASP2
## AC009950.1 AC009950.1
## SLC35F1 SLC35F1
## MEGF9 MEGF9
## MRPL53 MRPL53
## UBE3D UBE3D
## QSER1.2 QSER1
## VPS18.1 VPS18
## HYLS1 HYLS1
## ZNF83.2 ZNF83
## PDE4B.1 PDE4B
## SEMA4D SEMA4D
## ZMIZ2.1 ZMIZ2
## LIX1L-AS1 LIX1L-AS1
## KLF7.1 KLF7
## ZMAT1 ZMAT1
## VDR.1 VDR
## DNMT3A DNMT3A
## USP46.1 USP46
## UBFD1.1 UBFD1
## NUMB.1 NUMB
## NOL6 NOL6
## CD84.1 CD84
## SETD9 SETD9
## EPM2A.1 EPM2A
## MCM3AP MCM3AP
## KLHL28 KLHL28
## TLCD1.2 TLCD1
## FBXO43.2 FBXO43
## MANBA MANBA
## XXYLT1.1 XXYLT1
## GNB4 GNB4
## NREP.2 NREP
## AC123768.5 AC123768.5
## PPP1R16B.1 PPP1R16B
## LINC00920 LINC00920
## MTFR1.1 MTFR1
## PPP1R16A PPP1R16A
## ZNF460 ZNF460
## AC073254.1 AC073254.1
## NAB1.1 NAB1
## KLHL18.2 KLHL18
## KIAA0586.1 KIAA0586
## SLC66A1.1 SLC66A1
## RANBP10 RANBP10
## ZNF420 ZNF420
## TMEM94 TMEM94
## ANAPC2 ANAPC2
## POGZ.2 POGZ
## MAMSTR MAMSTR
## ICAM1 ICAM1
## AC009630.1 AC009630.1
## CBX8 CBX8
## TRBV24-1 TRBV24-1
## TYMSOS.1 TYMSOS
## LPAR6.1 LPAR6
## HMGN5 HMGN5
## FAM135A FAM135A
## TMSB4Y TMSB4Y
## SLC9A3R2 SLC9A3R2
## WDR41 WDR41
## HIST1H3C.2 HIST1H3C
## TMEM19 TMEM19
## CASP9 CASP9
## C1orf21.1 C1orf21
## UBAP1.1 UBAP1
## EIF3C EIF3C
## MAP1LC3A MAP1LC3A
## MLLT1.1 MLLT1
## ADAP1.1 ADAP1
## SLC30A4 SLC30A4
## GLMN.1 GLMN
## LRRC20.1 LRRC20
## PROSER3 PROSER3
## PIP5K1A.1 PIP5K1A
## TMCO3 TMCO3
## PIAS2.1 PIAS2
## SLC1A4.1 SLC1A4
## ARMC5 ARMC5
## MKS1 MKS1
## OPA3.1 OPA3
## KLF5.2 KLF5
## LRRC28.1 LRRC28
## LRATD2 LRATD2
## HABP4.1 HABP4
## ZBTB1 ZBTB1
## YAF2 YAF2
## PTPN9.1 PTPN9
## AC135457.1 AC135457.1
## DAAM1 DAAM1
## PIK3CG.1 PIK3CG
## C8orf58 C8orf58
## ANG.1 ANG
## C19orf57.2 C19orf57
## ZNF680 ZNF680
## GOLGA3 GOLGA3
## DICER1 DICER1
## TIPARP TIPARP
## NCOA6 NCOA6
## PRH1.1 PRH1
## ZNF687.1 ZNF687
## STK11IP STK11IP
## LRFN3 LRFN3
## TMEM229B TMEM229B
## TMSB15B.2 TMSB15B
## C17orf67 C17orf67
## NANP NANP
## OR4D1 OR4D1
## C17orf80 C17orf80
## PYROXD2 PYROXD2
## CARF CARF
## KIAA1109 KIAA1109
## RHEBL1.1 RHEBL1
## CCDC6.1 CCDC6
## NBPF15 NBPF15
## RBM26 RBM26
## DNHD1 DNHD1
## ZNF839 ZNF839
## AC145207.2.1 AC145207.2
## IGFBP6 IGFBP6
## CHRM3-AS2 CHRM3-AS2
## RCL1 RCL1
## SUFU.1 SUFU
## HOOK1.2 HOOK1
## ERI1.1 ERI1
## DIXDC1.2 DIXDC1
## PLAUR.1 PLAUR
## TGFBR3.1 TGFBR3
## NAA35 NAA35
## GTF3C1 GTF3C1
## FAM20B.1 FAM20B
## AC018816.1 AC018816.1
## DCAF6 DCAF6
## YLPM1.1 YLPM1
## EOGT.1 EOGT
## AC002467.1.1 AC002467.1
## OTUD4 OTUD4
## FLVCR1-DT FLVCR1-DT
## FLYWCH1 FLYWCH1
## C1orf54 C1orf54
## AJM1 AJM1
## PPP1R37 PPP1R37
## RNF38.1 RNF38
## TBCCD1.1 TBCCD1
## ZNF251 ZNF251
## FSD1 FSD1
## MBLAC2 MBLAC2
## IKZF3.2 IKZF3
## AC073529.1.2 AC073529.1
## EFCAB7 EFCAB7
## PVRIG.1 PVRIG
## PGM3.1 PGM3
## ABHD18 ABHD18
## ALS2 ALS2
## EPHX2.1 EPHX2
## FAM171A1.2 FAM171A1
## ALDH5A1 ALDH5A1
## RAB11FIP4 RAB11FIP4
## MSANTD2 MSANTD2
## STRBP.2 STRBP
## KIAA1841.1 KIAA1841
## PPP1R9B PPP1R9B
## KIF16B.2 KIF16B
## RAB11FIP3 RAB11FIP3
## RBBP9.1 RBBP9
## DGKE.1 DGKE
## C2CD3.1 C2CD3
## PAIP2B PAIP2B
## RGPD8 RGPD8
## NSMCE1-DT.1 NSMCE1-DT
## VSIG10 VSIG10
## AC114760.2 AC114760.2
## SULF2 SULF2
## RNF24.1 RNF24
## ZNF566 ZNF566
## C11orf49.1 C11orf49
## ALG11 ALG11
## RRAGB RRAGB
## AC067852.2 AC067852.2
## TRAF6 TRAF6
## NUP35.2 NUP35
## GGT1.1 GGT1.1
## HIST1H4D.1 HIST1H4D
## PCGF3 PCGF3
## MFSD13A MFSD13A
## SPATA20 SPATA20
## MAK MAK
## MESP1.1 MESP1
## ZNF121 ZNF121
## ARID3A ARID3A
## NBPF10 NBPF10
## KLHL6.1 KLHL6
## SMYD5.2 SMYD5
## AGBL5 AGBL5
## STRN STRN
## UBE2O UBE2O
## NAT1.1 NAT1
## XRRA1.1 XRRA1
## ERMARD ERMARD
## SP140 SP140
## SREBF2.1 SREBF2
## EME1.2 EME1
## PSRC1.2 PSRC1
## DCUN1D4 DCUN1D4
## AC011446.2 AC011446.2
## PPM1B PPM1B
## EXD2.1 EXD2
## CRYBG3.1 CRYBG3
## EDDM13.1 EDDM13
## C5orf58 C5orf58
## GPR19.1 GPR19
## FAR2.2 FAR2
## ZBTB21 ZBTB21
## PYROXD1.1 PYROXD1
## LMBR1 LMBR1
## TAF5 TAF5
## KDM4C KDM4C
## MTMR4.1 MTMR4
## EYA3.1 EYA3
## ZCCHC4 ZCCHC4
## ZBTB17 ZBTB17
## NDRG1 NDRG1
## FCRLB FCRLB
## AC005229.4 AC005229.4
## AC005695.2 AC005695.2
## B4GALT4 B4GALT4
## STK38.1 STK38
## PLEKHB1 PLEKHB1
## FGFR1OP.1 FGFR1OP
## DEPDC5 DEPDC5
## AC008083.2.1 AC008083.2
## AC074032.1 AC074032.1
## SYTL3.1 SYTL3
## STIM1.1 STIM1
## IFT88.1 IFT88
## TRIT1 TRIT1
## TPCN1.2 TPCN1
## DNAJC24.1 DNAJC24
## AL359220.1.1 AL359220.1
## ZNF829 ZNF829
## MORC2.2 MORC2
## CCNT2 CCNT2
## ROCK1.1 ROCK1
## AC073332.1 AC073332.1
## IPP IPP
## TRAV5 TRAV5
## ZNF333 ZNF333
## POLR1A.1 POLR1A
## AC099343.3 AC099343.3
## KLHL5.2 KLHL5
## FBXO32 FBXO32
## ZNF816 ZNF816
## AC159540.2 AC159540.2
## C17orf97 C17orf97
## AC008105.1 AC008105.1
## AC091982.3 AC091982.3
## CEP162.1 CEP162
## PAQR3.1 PAQR3
## DCBLD1 DCBLD1
## SRD5A1.1 SRD5A1
## ARL17A ARL17A
## ABHD2.1 ABHD2
## AP001267.1 AP001267.1
## TRIM25.1 TRIM25
## ATP8B1 ATP8B1
## AL137856.1 AL137856.1
## NOCT.1 NOCT
## ZNF492.2 ZNF492
## CHCHD6.2 CHCHD6
## IQCE.1 IQCE
## TRGV2.2 TRGV2
## PAK4 PAK4
## FAM149B1 FAM149B1
## SIN3A.2 SIN3A
## MTMR12 MTMR12
## ZNF354C ZNF354C
## AC010969.2 AC010969.2
## MACROD2 MACROD2
## CELF1 CELF1
## AC068888.1.1 AC068888.1
## ZNF585B ZNF585B
## ANKRD36B.1 ANKRD36B
## HPS4 HPS4
## ZNF791 ZNF791
## ELF3-AS1 ELF3-AS1
## F8A1 F8A1
## CACNB2 CACNB2
## TNFSF13B.2 TNFSF13B
## WNT5B WNT5B
## LMBRD2 LMBRD2
## FRMD4B.1 FRMD4B
## PISD PISD
## PPM1F.1 PPM1F
## KCNG2 KCNG2
## OAF OAF
## TRGC1.1 TRGC1
## ATG2A ATG2A
## COG6 COG6
## PTGIR.2 PTGIR
## DYRK3 DYRK3
## NRSN2-AS1 NRSN2-AS1
## LMTK2.1 LMTK2
## IL10RA.2 IL10RA
## CCDC102B CCDC102B
## TMEM25.2 TMEM25
## TMEM53 TMEM53
## AEBP2 AEBP2
## MFSD6 MFSD6
## PBX3.2 PBX3
## BTBD2 BTBD2
## MINK1.1 MINK1
## LPIN2 LPIN2
## AL445524.1 AL445524.1
## PIK3CA.1 PIK3CA
## ADAM12 ADAM12
## AC005037.1.1 AC005037.1
## MAVS MAVS
## STARD4.1 STARD4
## AC068724.3 AC068724.3
## PODXL2 PODXL2
## CXCL16 CXCL16
## RNF130.1 RNF130
## MBLAC1 MBLAC1
## TESMIN.1 TESMIN
## AL139807.1.1 AL139807.1
## DNAJC27-AS1 DNAJC27-AS1
## POLM POLM
## GCLC GCLC
## FBXO3 FBXO3
## GPR146 GPR146
## EPHA4.2 EPHA4
## TRBV30.2 TRBV30
## NPHP4.2 NPHP4
## PHOSPHO2 PHOSPHO2
## TULP4 TULP4
## DCAF1.2 DCAF1
## BICD1.1 BICD1
## SEMA4B SEMA4B
## AP000640.2 AP000640.2
## EPC1.1 EPC1
## WDR5B WDR5B
## SH3PXD2A.1 SH3PXD2A
## TMPRSS6 TMPRSS6
## TNFRSF14-AS1 TNFRSF14-AS1
## C12orf4.1 C12orf4
## SEPSECS-AS1 SEPSECS-AS1
## SPON1-AS1 SPON1-AS1
## EARS2.1 EARS2
## AC092919.1 AC092919.1
## ISM1.1 ISM1
## AC087276.2 AC087276.2
## LINC01806 LINC01806
## AC016065.1 AC016065.1
## MAPK1.2 MAPK1
## LATS1 LATS1
## PTCD2 PTCD2
## ZNF548 ZNF548
## TRUB1.1 TRUB1
## IPCEF1 IPCEF1
## C5orf24 C5orf24
## DNM2 DNM2
## SLC12A7.1 SLC12A7
## SOS2 SOS2
## PACSIN2.1 PACSIN2
## ATN1.2 ATN1
## MKRN2OS MKRN2OS
## PLEKHG4 PLEKHG4
## GTPBP2.1 GTPBP2
## METRNL.1 METRNL
## PAXIP1-AS1 PAXIP1-AS1
## FAM78A FAM78A
## JAK3.1 JAK3
## ZFP69 ZFP69
## RAB30 RAB30
## SLC30A6.1 SLC30A6
## WARS2-AS1 WARS2-AS1
## KDM4A KDM4A
## ZNF583 ZNF583
## TSTD3 TSTD3
## SPATA2L SPATA2L
## MARK4 MARK4
## CNKSR2.2 CNKSR2
## ENO3 ENO3
## FADS2.1 FADS2
## OSBPL8 OSBPL8
## CCR1.2 CCR1
## DBT DBT
## AC118549.1 AC118549.1
## FCRL6.1 FCRL6
## ZNF850 ZNF850
## TNIP3.1 TNIP3
## SRSF11 SRSF11
## AC104316.1 AC104316.1
## RNF144A-AS1.2 RNF144A-AS1
## ADIPOR2 ADIPOR2
## RNF31.1 RNF31
## LINC01970 LINC01970
## PRR12 PRR12
## SLC9A6.1 SLC9A6
## ST3GAL5.2 ST3GAL5
## ABHD8.1 ABHD8
## AC009690.2 AC009690.2
## HIP1R HIP1R
## ARHGAP11B.2 ARHGAP11B
## ATP2A2.2 ATP2A2
## GPR35.1 GPR35
## SYCP2.1 SYCP2
## CCDC163 CCDC163
## C1orf53 C1orf53
## TCEAL3 TCEAL3
## ENTPD7 ENTPD7
## SLC44A1.2 SLC44A1
## TLK2.1 TLK2
## RP2 RP2
## VPS13C VPS13C
## ZNF324B ZNF324B
## SOCS7 SOCS7
## CEP192.2 CEP192
## SLC31A2.1 SLC31A2
## ZNF484 ZNF484
## EAF2.2 EAF2
## MDFIC.1 MDFIC
## MIR222HG.1 MIR222HG
## LINC00649.1 LINC00649
## AC087294.1 AC087294.1
## SYNRG.1 SYNRG
## MKNK1.2 MKNK1
## ASH1L ASH1L
## MLYCD MLYCD
## UBXN7 UBXN7
## DCP1A DCP1A
## ZNF736.2 ZNF736
## SNX32 SNX32
## DMC1.2 DMC1
## PEX11A PEX11A
## ZNF335 ZNF335
## CNNM3.1 CNNM3
## FEZ2.1 FEZ2
## ANGEL2 ANGEL2
## PDK3.1 PDK3
## AC010976.1.1 AC010976.1
## POLR3G POLR3G
## TOLLIP-AS1.2 TOLLIP-AS1
## ZNF219 ZNF219
## AC004158.1 AC004158.1
## CLSTN1 CLSTN1
## AP000873.2 AP000873.2
## KAT14.1 KAT14
## CDKL1.1 CDKL1
## ZNF514 ZNF514
## HSPA14 HSPA14
## BBS2 BBS2
## NICN1 NICN1
## IKZF4.1 IKZF4
## TSNARE1.1 TSNARE1
## MAP1A MAP1A
## AC058791.1 AC058791.1
## AC009126.1 AC009126.1
## SLC26A4-AS1.1 SLC26A4-AS1
## UFSP1 UFSP1
## MAN1B1-DT MAN1B1-DT
## AC015849.1.1 AC015849.1
## GPR89B GPR89B
## ADNP2.2 ADNP2
## JHY.1 JHY
## KHNYN KHNYN
## ZMIZ1 ZMIZ1
## CENPO.2 CENPO
## STAM.1 STAM
## AC092652.1 AC092652.1
## SMG7.1 SMG7
## DUSP5.1 DUSP5
## BTBD7 BTBD7
## MXD1 MXD1
## IQCH IQCH
## RAD9B RAD9B
## MYO9B MYO9B
## GGA3 GGA3
## SLC16A7.1 SLC16A7
## MFSD9 MFSD9
## IL6R.2 IL6R
## TTC38.1 TTC38
## NDN.2 NDN
## DERPC DERPC
## COPZ2 COPZ2
## AC021092.1 AC021092.1
## AL162253.2 AL162253.2
## HEATR5A HEATR5A
## BMERB1 BMERB1
## PAXBP1.2 PAXBP1
## POLR2J2 POLR2J2
## MTERF2 MTERF2
## LNX2 LNX2
## NIBAN2 NIBAN2
## TAF5L TAF5L
## ZNF707 ZNF707
## TAF4B.2 TAF4B
## KIF27 KIF27
## ARHGAP35.1 ARHGAP35
## C2orf81 C2orf81
## PDCD1LG2 PDCD1LG2
## INTS6-AS1 INTS6-AS1
## AC022916.1 AC022916.1
## RAP1GAP2.1 RAP1GAP2
## NAA25.2 NAA25
## CEP164 CEP164
## ATP7A ATP7A
## SPATA5.2 SPATA5
## TDRKH TDRKH
## ZNF180 ZNF180
## IFI44.2 IFI44
## CLOCK CLOCK
## TAF1A TAF1A
## ZNF90.2 ZNF90
## AL121944.1 AL121944.1
## TCAF2 TCAF2
## BX664615.2 BX664615.2
## MTF1 MTF1
## B3GAT2 B3GAT2
## AL691403.1.1 AL691403.1
## DIAPH2.2 DIAPH2
## PIGV PIGV
## ZNFX1 ZNFX1
## PPME1.1 PPME1
## TSGA10 TSGA10
## ERICH6-AS1 ERICH6-AS1
## TRAV1-2.1 TRAV1-2
## CEP70 CEP70
## GLCE.2 GLCE
## TARSL2 TARSL2
## TRDV1 TRDV1
## SYNGR3 SYNGR3
## KDM3B.1 KDM3B
## AP002381.2 AP002381.2
## Z93930.2 Z93930.2
## RPRD1A.1 RPRD1A
## KTI12 KTI12
## IPPK IPPK
## POGLUT3 POGLUT3
## MDM4 MDM4
## GPR137B.1 GPR137B
## FAM131A FAM131A
## FKBP1B.1 FKBP1B
## POC1B.2 POC1B
## GALNT11.1 GALNT11
## TRMT10A TRMT10A
## THBS4 THBS4
## AC073569.2 AC073569.2
## ZNF784 ZNF784
## WDR91 WDR91
## CYB5D2 CYB5D2
## PUS10 PUS10
## B3GALNT2.1 B3GALNT2
## UTY UTY
## ATXN7L1.1 ATXN7L1
## SLC2A1 SLC2A1
## APH1B APH1B
## CLIC4.2 CLIC4
## AC010260.1.1 AC010260.1
## MNT.1 MNT
## NOTCH2NLC NOTCH2NLC
## IKBKB.2 IKBKB
## NACC2 NACC2
## PLCB3.1 PLCB3
## LINC02802 LINC02802
## AMIGO1.1 AMIGO1
## MTUS1 MTUS1
## USP12-AS1 USP12-AS1
## PLLP PLLP
## SLC35A3.1 SLC35A3
## NIPA1 NIPA1
## AC245060.5 AC245060.5
## PIGA PIGA
## AP002812.2 AP002812.2
## TRAM2-AS1 TRAM2-AS1
## FGF14-AS2 FGF14-AS2
## LKAAEAR1.2 LKAAEAR1
## SLC29A2 SLC29A2
## AC010149.1 AC010149.1
## LINC01765.1 LINC01765
## TTC21B-AS1 TTC21B-AS1
## UBE2W UBE2W
## TRIM68 TRIM68
## RNF43 RNF43
## AL031728.1 AL031728.1
## TRBV6-2 TRBV6-2
## DDX43 DDX43
## VWA5A VWA5A
## EXOC3-AS1 EXOC3-AS1
## AC026202.2 AC026202.2
## TNFRSF10D TNFRSF10D
## VPS52 VPS52
## RPTOR RPTOR
## MRTFB.1 MRTFB
## NLRP1.1 NLRP1
## SNRK.1 SNRK
## AP2B1 AP2B1
## ELMOD3.1 ELMOD3
## ZFY ZFY
## AC005670.3.1 AC005670.3
## RUNDC1 RUNDC1
## TRAV24.1 TRAV24
## GTDC1 GTDC1
## ABR.1 ABR
## FBXL22 FBXL22
## GATD3A GATD3A
## MLF1 MLF1
## TIPARP-AS1 TIPARP-AS1
## PIGL PIGL
## AC027644.3 AC027644.3
## LINC01089.1 LINC01089
## CFLAR.2 CFLAR
## NIPBL-DT NIPBL-DT
## HIST1H2AJ.1 HIST1H2AJ
## ZNF382 ZNF382
## VGLL4 VGLL4
## SLFN13 SLFN13
## AC004596.1 AC004596.1
## RASGRP3.1 RASGRP3
## EHBP1L1.1 EHBP1L1
## FAM92A FAM92A
## RLN2.1 RLN2
## RBM26-AS1 RBM26-AS1
## ZNF284 ZNF284
## BTG3-AS1 BTG3-AS1
## CD274 CD274
## ZNF91 ZNF91
## DST.2 DST
## AC036176.1.1 AC036176.1
## ZSCAN18 ZSCAN18
## NEK3 NEK3
## BBS4 BBS4
## BCAS3 BCAS3
## ACVR2A.1 ACVR2A
## PPARG PPARG
## TTC33.1 TTC33
## APLF.1 APLF
## PIAS1 PIAS1
## MRNIP MRNIP
## RTTN.2 RTTN
## STK35.2 STK35
## METTL22 METTL22
## CYFIP1.2 CYFIP1
## AP4S1 AP4S1
## KLF9 KLF9
## AP001318.2 AP001318.2
## KDM7A-DT KDM7A-DT
## CXorf21 CXorf21
## FNBP4 FNBP4
## ADCK5 ADCK5
## P3H4 P3H4
## PINLYP PINLYP
## ZC3H12D ZC3H12D
## ACSF3.1 ACSF3
## AFAP1.2 AFAP1
## TIMD4.1 TIMD4
## FXYD1 FXYD1
## AC097382.2 AC097382.2
## LRRC8C.1 LRRC8C
## TIRAP TIRAP
## USP3 USP3
## DHX57 DHX57
## CEP250 CEP250
## SAT1 SAT1
## C19orf38.2 C19orf38
## AC011447.3.2 AC011447.3
## ETV3 ETV3
## MIR3142HG.1 MIR3142HG
## CBWD1 CBWD1
## ZFAND4.1 ZFAND4
## TRGV8.1 TRGV8
## ZC3H10 ZC3H10
## THRA THRA
## AL645728.1.1 AL645728.1
## SUN1.2 SUN1
## HINFP HINFP
## ACSL4 ACSL4
## TUT7.1 TUT7
## ACSF2 ACSF2
## KPTN.1 KPTN
## HIST1H4F HIST1H4F
## FRMD5.2 FRMD5
## LRRC75A LRRC75A
## BDNF-AS BDNF-AS
## LINC01260 LINC01260
## POMGNT2 POMGNT2
## TMEM260 TMEM260
## KIF5C.1 KIF5C
## MTHFD2L MTHFD2L
## ZNF737.1 ZNF737
## SIRPG-AS1 SIRPG-AS1
## DDX31 DDX31
## LGALS8-AS1 LGALS8-AS1
## TCTN1 TCTN1
## HYPK HYPK
## TRBV12-4.1 TRBV12-4
## B3GNT10 B3GNT10
## RBL2.2 RBL2
## ABCC4.1 ABCC4
## NSD1 NSD1
## MAFF MAFF
## TRBV4-1.1 TRBV4-1
## SIRT1 SIRT1
## GCNA GCNA
## TBC1D25 TBC1D25
## ZNF318 ZNF318
## TATDN2 TATDN2
## MYO5A.1 MYO5A
## HOMER2.1 HOMER2
## SMAD5 SMAD5
## TLN1.1 TLN1
## TMEM39A TMEM39A
## TOP3B TOP3B
## NKIRAS1 NKIRAS1
## CUL1.2 CUL1
## AC020913.1.1 AC020913.1
## BBOF1 BBOF1
## LINC02580 LINC02580
## LINC00092 LINC00092
## AL139099.1 AL139099.1
## LINC00891.1 LINC00891
## AK9 AK9
## R3HCC1L R3HCC1L
## PHF8.1 PHF8
## L3MBTL3 L3MBTL3
## ZNF551 ZNF551
## TRAF3IP2-AS1 TRAF3IP2-AS1
## GATA3-AS1.2 GATA3-AS1
## QKI.1 QKI
## SSH2.1 SSH2
## AC008124.1 AC008124.1
## SLC39A13 SLC39A13
## ARHGAP26.1 ARHGAP26
## ZNF510 ZNF510
## CD79A CD79A
## AL121787.1 AL121787.1
## AC090061.1 AC090061.1
## TBXA2R.1 TBXA2R
## PHETA2 PHETA2
## FNTB.1 FNTB
## CHST10 CHST10
## PIGZ.1 PIGZ
## ELK3 ELK3
## MAPK8 MAPK8
## ANKS6 ANKS6
## MAP3K14-AS1 MAP3K14-AS1
## PKN3.2 PKN3
## SRCAP SRCAP
## RTL10 RTL10
## PDE8A.2 PDE8A
## ZNF43.2 ZNF43
## MYSM1 MYSM1
## AC108134.3 AC108134.3
## LINC00662 LINC00662
## AC018647.3 AC018647.3
## C16orf71 C16orf71
## ITGAM.2 ITGAM
## LDLR.2 LDLR
## HSPBAP1 HSPBAP1
## CDC37L1-DT CDC37L1-DT
## CCDC91 CCDC91
## PTPRE.1 PTPRE
## ZNF468 ZNF468
## AARS2 AARS2
## FHOD1 FHOD1
## SLC48A1 SLC48A1
## HCG11 HCG11
## SMC1B.1 SMC1B
## WAKMAR2.1 WAKMAR2
## AL355574.1 AL355574.1
## TIGD7 TIGD7
## CCDC7 CCDC7
## ARMH4 ARMH4
## PXYLP1 PXYLP1
## ZNF768 ZNF768
## SLC5A10 SLC5A10
## PSMA8 PSMA8
## ACTR5 ACTR5
## RNF111 RNF111
## SPIN3 SPIN3
## C18orf54.1 C18orf54
## SLC7A5.1 SLC7A5
## CSGALNACT1.2 CSGALNACT1
## MMP24 MMP24
## MBOAT1.1 MBOAT1
## ZCWPW2 ZCWPW2
## ACTR3B ACTR3B
## AL135925.1 AL135925.1
## PEX7.1 PEX7
## NME7.1 NME7
## VPS50 VPS50
## TCF25 TCF25
## CAMTA2.1 CAMTA2
## TRBV3-1 TRBV3-1
## FGL2 FGL2
## ERI2.2 ERI2
## NFE2L1.1 NFE2L1
## Z93241.1.1 Z93241.1
## DZANK1 DZANK1
## PLA2G15.1 PLA2G15
## NLK NLK
## PPARD.1 PPARD
## HDAC8 HDAC8
## IRF4.1 IRF4
## ZNF354A ZNF354A
## SLC29A3 SLC29A3
## CD160.1 CD160
## CES1.1 CES1
## AC080112.1 AC080112.1
## PDE9A PDE9A
## FKBP14.1 FKBP14
## TRAPPC8 TRAPPC8
## ZNF786 ZNF786
## PTGDR.1 PTGDR
## AC016727.1 AC016727.1
## MTERF1 MTERF1
## AC073342.1 AC073342.1
## SLC3A1 SLC3A1
## PARS2 PARS2
## AC107081.2 AC107081.2
## LINC00239.1 LINC00239
## CBX2.1 CBX2
## SIGLEC10 SIGLEC10
## LENG9 LENG9
## GRAMD4.1 GRAMD4
## SLC25A44 SLC25A44
## GANC GANC
## CEACAM1.1 CEACAM1
## CYP2U1 CYP2U1
## LYSMD4 LYSMD4
## TBCD.2 TBCD
## ST8SIA6.1 ST8SIA6
## METTL25 METTL25
## SLC2A8.1 SLC2A8
## HDAC4-AS1 HDAC4-AS1
## ZNF669 ZNF669
## KCNK12.1 KCNK12
## BBS12 BBS12
## MYOM2 MYOM2
## EXOC6B.2 EXOC6B
## DHX35 DHX35
## BAIAP2L1 BAIAP2L1
## DNAJC18 DNAJC18
## ACACA.1 ACACA
## ZNF441 ZNF441
## MLLT3.1 MLLT3
## LPIN1 LPIN1
## ETNK1 ETNK1
## AC018797.3 AC018797.3
## MIATNB MIATNB
## METTL7A.1 METTL7A
## NDRG2.1 NDRG2
## CTDSPL.2 CTDSPL
## INO80 INO80
## CD44 CD44
## ARHGEF9 ARHGEF9
## YTHDF3 YTHDF3
## RBM44 RBM44
## CBWD3.1 CBWD3
## CALCB.1 CALCB
## EVI5L EVI5L
## EEF2K EEF2K
## DHTKD1.1 DHTKD1
## ECM2.2 ECM2
## OMD.1 OMD
## ABCA3.1 ABCA3
## FAM185A FAM185A
## SPON1.1 SPON1
## JAKMIP1 JAKMIP1
## TICAM2 TICAM2
## CARD9 CARD9
## CCDC15-DT CCDC15-DT
## EP400.1 EP400
## ZNF25 ZNF25
## TDRD3 TDRD3
## DDX17 DDX17
## LIF.2 LIF
## SLC31A1.1 SLC31A1
## TSC2 TSC2
## ASNS ASNS
## AC008467.1 AC008467.1
## ITGA1.2 ITGA1
## GRAP2.1 GRAP2
## ZNF623 ZNF623
## MLLT6.1 MLLT6
## TTN-AS1.1 TTN-AS1
## RASA2.2 RASA2
## PIK3C3 PIK3C3
## AC027097.1 AC027097.1
## FZD6.1 FZD6
## RAB11FIP2 RAB11FIP2
## DACT1.2 DACT1
## SYCP3.1 SYCP3
## LINC01547 LINC01547
## KLF13.2 KLF13
## MTSS1 MTSS1
## TMEM168 TMEM168
## AC022075.1.2 AC022075.1
## EXOC2 EXOC2
## CD8B2.1 CD8B2
## IL4.1 IL4
## AC072022.2 AC072022.2
## RNASE6.1 RNASE6
## RNF32 RNF32
## MMP25.1 MMP25
## UNC119B UNC119B
## SEL1L3.2 SEL1L3
## ILDR2.2 ILDR2
## NEURL1.1 NEURL1
## PGM2L1 PGM2L1
## AC021188.1.1 AC021188.1
## PTPN4.2 PTPN4
## AGTPBP1 AGTPBP1
## ENPP4.1 ENPP4
## TP73.2 TP73
## LRP5 LRP5
## AC023509.2 AC023509.2
## ANKRD52.1 ANKRD52
## NNT.1 NNT
## AC064807.1.1 AC064807.1
## PPP1R12C PPP1R12C
## ACACB ACACB
FeaturePlot_scCustom(seu, features = "CD3E")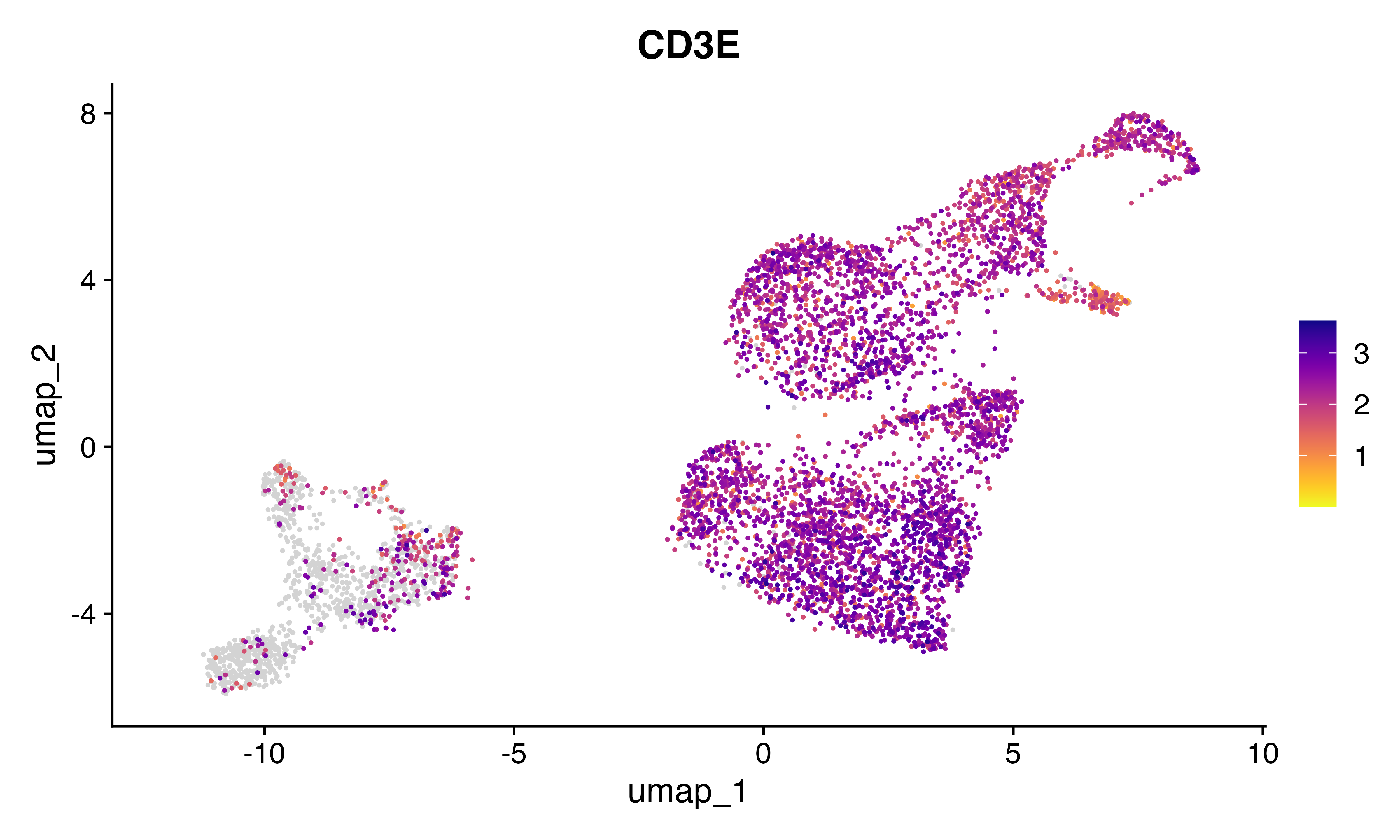
mito.genes <- grep(pattern = "^MT-", x = rownames(x = seu), value = TRUE)
seu$percent.mito <- Matrix::colSums(GetAssayData(seu[mito.genes, ]))/Matrix::colSums(GetAssayData(seu))
FeaturePlot_scCustom(seu, features = "percent.mito")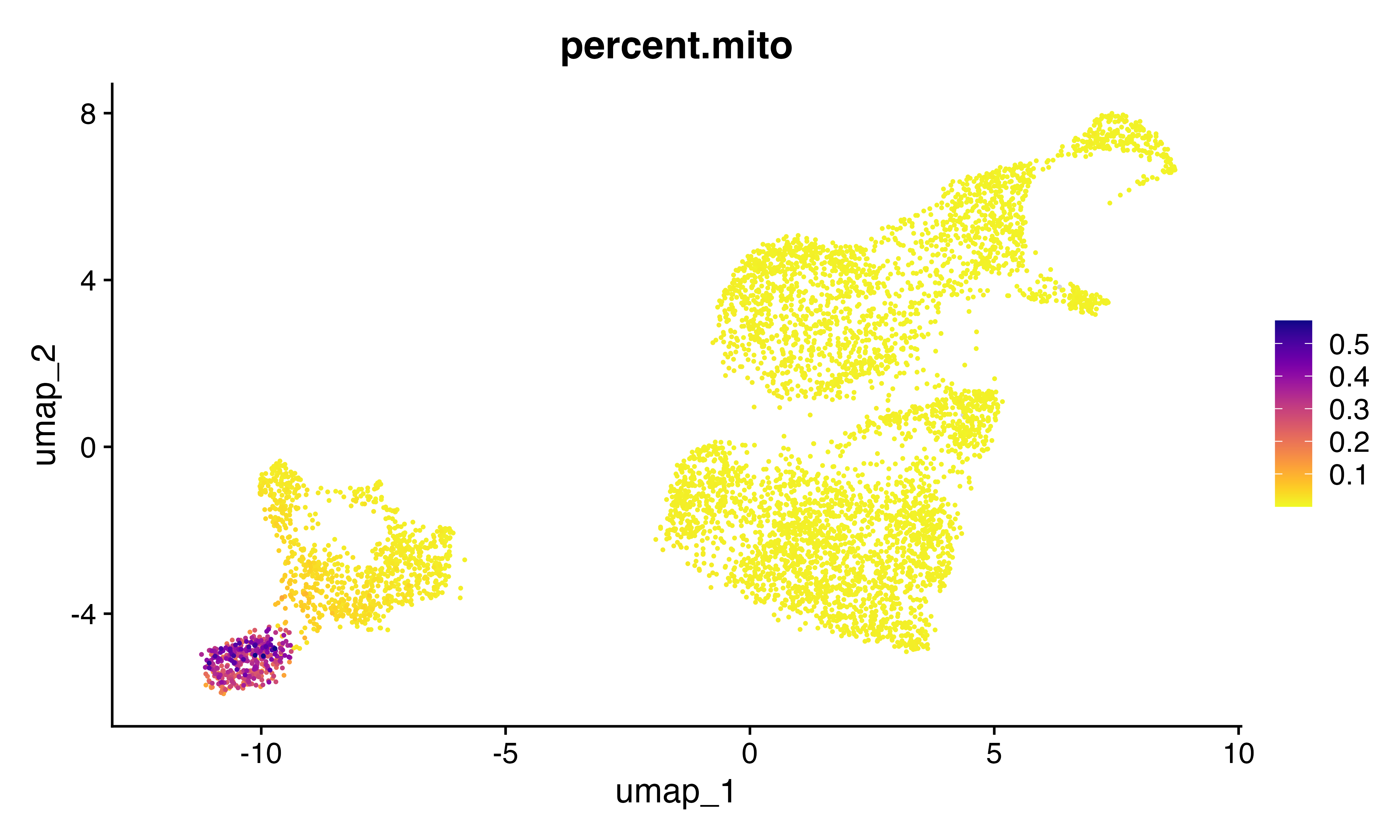
Much better. We see the data is more simple and we have more readily distinguishable clusters. ## Appendix
## R version 4.3.1 (2023-06-16)
## Platform: x86_64-apple-darwin20 (64-bit)
## Running under: macOS Ventura 13.6.3
##
## Matrix products: default
## BLAS: /Library/Frameworks/R.framework/Versions/4.3-x86_64/Resources/lib/libRblas.0.dylib
## LAPACK: /Library/Frameworks/R.framework/Versions/4.3-x86_64/Resources/lib/libRlapack.dylib; LAPACK version 3.11.0
##
## locale:
## [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
##
## time zone: America/Los_Angeles
## tzcode source: internal
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] magrittr_2.0.3 ggplot2_3.4.4 scrubletR_0.2.0 scCustomize_2.0.1
## [5] Seurat_5.0.1.9004 SeuratObject_5.0.1 sp_2.1-3 viewmastR_0.2.1
##
## loaded via a namespace (and not attached):
## [1] fs_1.6.3 matrixStats_1.2.0
## [3] spatstat.sparse_3.0-3 bitops_1.0-7
## [5] RcppMsgPack_0.2.3 lubridate_1.9.3
## [7] httr_1.4.7 RColorBrewer_1.1-3
## [9] doParallel_1.0.17 tools_4.3.1
## [11] sctransform_0.4.1 backports_1.4.1
## [13] utf8_1.2.4 R6_2.5.1
## [15] lazyeval_0.2.2 uwot_0.1.16
## [17] GetoptLong_1.0.5 withr_3.0.0
## [19] gridExtra_2.3 progressr_0.14.0
## [21] cli_3.6.2 Biobase_2.60.0
## [23] textshaping_0.3.7 Cairo_1.6-2
## [25] spatstat.explore_3.2-6 fastDummies_1.7.3
## [27] labeling_0.4.3 sass_0.4.8
## [29] spatstat.data_3.0-4 proxy_0.4-27
## [31] ggridges_0.5.6 pbapply_1.7-2
## [33] pkgdown_2.0.7 systemfonts_1.0.5
## [35] foreign_0.8-86 parallelly_1.36.0
## [37] limma_3.56.2 rstudioapi_0.15.0
## [39] generics_0.1.3 shape_1.4.6
## [41] ica_1.0-3 spatstat.random_3.2-2
## [43] dplyr_1.1.4 Matrix_1.6-5
## [45] ggbeeswarm_0.7.2 fansi_1.0.6
## [47] S4Vectors_0.38.2 abind_1.4-5
## [49] lifecycle_1.0.4 yaml_2.3.8
## [51] snakecase_0.11.1 SummarizedExperiment_1.30.2
## [53] recipes_1.0.9 Rtsne_0.17
## [55] paletteer_1.6.0 grid_4.3.1
## [57] promises_1.2.1 crayon_1.5.2
## [59] miniUI_0.1.1.1 lattice_0.22-5
## [61] cowplot_1.1.3 pillar_1.9.0
## [63] knitr_1.45 ComplexHeatmap_2.16.0
## [65] GenomicRanges_1.52.1 rjson_0.2.21
## [67] boot_1.3-28.1 future.apply_1.11.1
## [69] codetools_0.2-19 leiden_0.4.3.1
## [71] glue_1.7.0 data.table_1.15.0
## [73] vctrs_0.6.5 png_0.1-8
## [75] spam_2.10-0 gtable_0.3.4
## [77] rematch2_2.1.2 assertthat_0.2.1
## [79] cachem_1.0.8 gower_1.0.1
## [81] xfun_0.41 S4Arrays_1.2.0
## [83] mime_0.12 prodlim_2023.08.28
## [85] survival_3.5-7 timeDate_4032.109
## [87] SingleCellExperiment_1.22.0 iterators_1.0.14
## [89] pbmcapply_1.5.1 hardhat_1.3.0
## [91] lava_1.7.3 ellipsis_0.3.2
## [93] fitdistrplus_1.1-11 ROCR_1.0-11
## [95] ipred_0.9-14 nlme_3.1-164
## [97] bit64_4.0.5 RcppAnnoy_0.0.22
## [99] GenomeInfoDb_1.36.4 bslib_0.6.1
## [101] irlba_2.3.5.1 vipor_0.4.7
## [103] KernSmooth_2.23-22 rpart_4.1.23
## [105] colorspace_2.1-0 BiocGenerics_0.46.0
## [107] Hmisc_5.1-1 nnet_7.3-19
## [109] ggrastr_1.0.2 tidyselect_1.2.0
## [111] bit_4.0.5 compiler_4.3.1
## [113] htmlTable_2.4.2 hdf5r_1.3.9
## [115] desc_1.4.3 DelayedArray_0.26.7
## [117] plotly_4.10.4 checkmate_2.3.1
## [119] scales_1.3.0 lmtest_0.9-40
## [121] stringr_1.5.1 digest_0.6.34
## [123] goftest_1.2-3 presto_1.0.0
## [125] spatstat.utils_3.0-4 minqa_1.2.6
## [127] rmarkdown_2.25 XVector_0.40.0
## [129] htmltools_0.5.7 pkgconfig_2.0.3
## [131] base64enc_0.1-3 lme4_1.1-35.1
## [133] sparseMatrixStats_1.12.2 MatrixGenerics_1.12.3
## [135] highr_0.10 fastmap_1.1.1
## [137] rlang_1.1.3 GlobalOptions_0.1.2
## [139] htmlwidgets_1.6.4 shiny_1.8.0
## [141] DelayedMatrixStats_1.22.6 farver_2.1.1
## [143] jquerylib_0.1.4 zoo_1.8-12
## [145] jsonlite_1.8.8 ModelMetrics_1.2.2.2
## [147] RCurl_1.98-1.14 Formula_1.2-5
## [149] GenomeInfoDbData_1.2.10 dotCall64_1.1-1
## [151] patchwork_1.2.0 munsell_0.5.0
## [153] Rcpp_1.0.12 reticulate_1.35.0
## [155] stringi_1.8.3 pROC_1.18.5
## [157] zlibbioc_1.46.0 MASS_7.3-60.0.1
## [159] plyr_1.8.9 parallel_4.3.1
## [161] listenv_0.9.1 ggrepel_0.9.5
## [163] forcats_1.0.0 deldir_2.0-2
## [165] splines_4.3.1 tensor_1.5
## [167] circlize_0.4.15 igraph_2.0.1.1
## [169] spatstat.geom_3.2-8 RcppHNSW_0.6.0
## [171] reshape2_1.4.4 stats4_4.3.1
## [173] evaluate_0.23 ggprism_1.0.4
## [175] nloptr_2.0.3 foreach_1.5.2
## [177] httpuv_1.6.14 RANN_2.6.1
## [179] tidyr_1.3.1 purrr_1.0.2
## [181] polyclip_1.10-6 future_1.33.1
## [183] clue_0.3-65 scattermore_1.2
## [185] janitor_2.2.0 xtable_1.8-4
## [187] monocle3_1.4.3 e1071_1.7-14
## [189] RSpectra_0.16-1 later_1.3.2
## [191] viridisLite_0.4.2 class_7.3-22
## [193] ragg_1.2.7 tibble_3.2.1
## [195] memoise_2.0.1 beeswarm_0.4.0
## [197] IRanges_2.34.1 cluster_2.1.6
## [199] timechange_0.3.0 globals_0.16.2
## [201] caret_6.0-94
getwd()## [1] "/Users/sfurlan/develop/scrubletR/vignettes"